- Title
-
HNF factors form a network to regulate liver-enriched genes in zebrafish
- Authors
- Cheng, W., Guo, L., Zhang, Z., Soo, H.M., Wen, C., Wu, W., and Peng, J.
- Source
- Full text @ Dev. Biol.
|
Classification of liver-enriched genes in zebrafish. RNA samples were extracted from the dissected liver and liver-free body, respectively. In panel A, RNA samples from 3 dpf embryos were also included. Probes were derived from EST clones or gene fragments cloned in plasmid vector via RT-PCR using gene-specific primers (Table 1; Supplementary Table 4). RNA hybridization blots were arranged according to the descending ratio of expression levels of liver vs. liver-free body (as indicated in the bracket). We arbitrarily grouped this set of genes according to the ratio of expression level. Genes with ratios more than 6.9 are considered as liver-specific genes, ratios between 6.9 and 2 as highly-liver-enriched genes and ratios less than 2 as moderately-liver-enriched genes. Clone identification is shown on the right side. Hybridization intensity was quantified using software Molecular Analyst (Bio-rad). B, liver-free body; L, liver; E, 3 dpf embryos. EXPRESSION / LABELING:
|
|
Identification of 69 genes enriched in the embryonic liver. Probes were derived from all 129 adult liver-enriched genes and were used to perform whole-mount in situ hybridization on 3 dpf, 4 dpf and 5 dpf embryos, respectively. (A) Expression of 56 genes is predominantly enriched in the liver but not in other digestive organs. (B) Expression of six genes is enriched in the liver and intestine but not in the pancreas. (C) Expression of six genes is enriched in all three major digestive organs. Clone identification is shown on the right corner. The liver is indicated by black arrow, the intestine by red arrow and the pancreas by green arrow. EXPRESSION / LABELING:
|
|
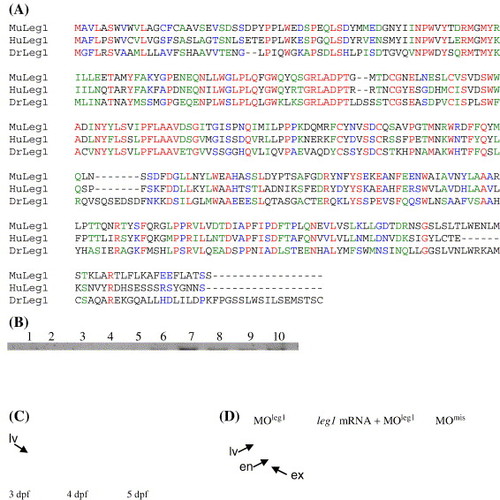
Leg1 functions as a novel factor involved in controlling liver expansion growth. (A) Amino acid sequence alignment of Leg1 (DrLeg1, CT737258) with its homologues in human (HuLeg1, gi60502446) and mouse (MuLeg1) gi12844359). (B) RNA gel blot hybridization revealed that leg1 is undetectable before 17 hpf (lanes 1?5: 1-cell-stage, 5 hpf, 9 hpf, 12 hpf and 17 hpf, respectively). leg1 starts to express at 24 hpf and is maintained at relative high level through the rest of embryonic stages (lanes 6?10: 1 dpf, 2 dpf, 3 dpf, 4 dpf and 5 dpf, respectively). (C) Whole-mount in situ hybridization using a leg1 probe revealed that leg1 is enriched in the embryonic liver. (D) MOleg1 morphants conferred a smaller liver phenotype (35 out of 42 morphants examined; representative embryo shown on the left) when compared with that of a control MOmis morphant (embryo on the right). Co-injection of the leg1 mRNA with MOleg1 restored the morphant phenotype to normal (embryo in the middle; 26 out 38 embryos examined). Embryos were stained with triple markers: a secreted immunoglobulin 4 probe as the liver (lv) marker, an insulin probe as the endocrine pancreas (en) marker and a trypsin probe as the exocrine pancreas (ex) marker. |
|
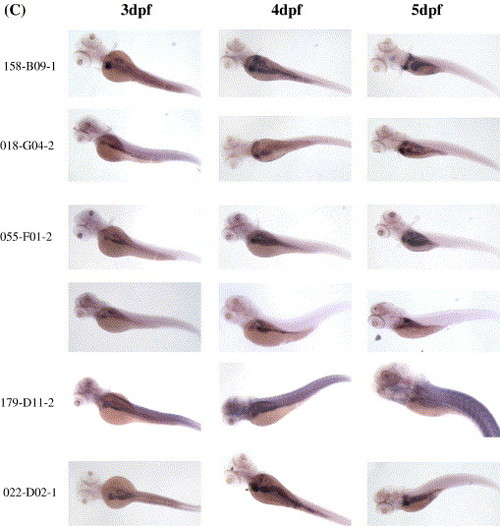
Whole-mount in situ hybridization was performed on 3 dpf, 4 dpf and 5 dpf embryos to identify genes enriched in the embryonic liver. Clone identifications (clone ID) are provided on the left side of each panel. (A) 56 genes enriched mainly in the liver. (B) 6 genes enriched in the liver and gut but not in the pancreas. (C) 6 genes enriched in the liver, pancreas and gut. |
|
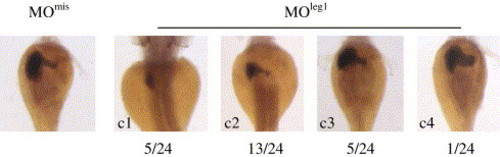
Majority of morphants injected with specific leg1 morpholino MOleg1 caused smaller liver. Four categories (c1?c4) of morphants were observed and the first two categories (c1 plus c2, 18 out of 24 embryos examined) displayed obvious smaller liver. EXPRESSION / LABELING:
|
Reprinted from Developmental Biology, 294(2), Cheng, W., Guo, L., Zhang, Z., Soo, H.M., Wen, C., Wu, W., and Peng, J., HNF factors form a network to regulate liver-enriched genes in zebrafish, 482-496, Copyright (2006) with permission from Elsevier. Full text @ Dev. Biol.