- Title
-
Sec13 safeguards the integrity of the endoplasmic reticulum and organogenesis of the digestive system in zebrafish
- Authors
- Niu, X., Gao, C., Jan Lo, L., Luo, Y., Meng, C., Hong, J., Hong, W., and Peng, J.
- Source
- Full text @ Dev. Biol.
|
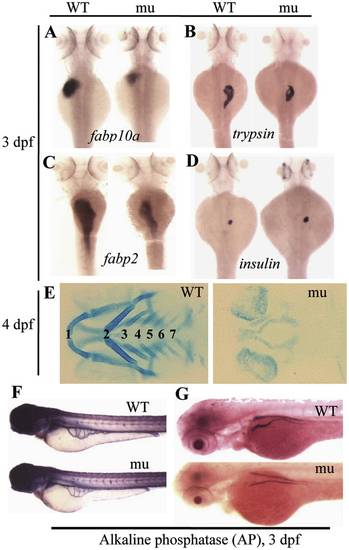
The sq198 mutation confers hypoplastic digestive organs and malformed skeletal cartilage. (A?D) WISH using markers specifically for the liver (fabp10a) (A), exocrine pancreas (trypsin) (B), intestine (fabp2) (C), and endocrine pancreas (insulin) (D) on 3 dpf wildtype (WT) and mutant (mu) embryos. (E) Alcian blue staining showing the branchial arches 1?7 in WT and sq198 (mu) at 4 dpf. (F,G) Histochemical staining of alkaline phosphatase (AP) did not reveal obvious difference between 3 dpf wildtype (WT) and mutant (mu) blood vessels (F) whilst the head region of the pronephric ducts appears thinner in the mutant (G). EXPRESSION / LABELING:
PHENOTYPE:
|
|
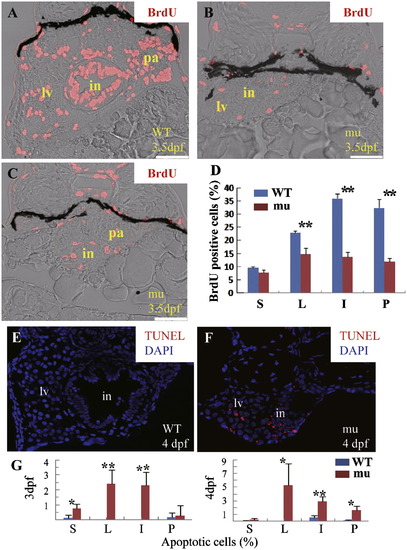
The sq198 mutation causes cell cycle arrest and cell apoptosis in the digestive organs. (A?D) BrdU incorporation assay showed that, at 3.5 dpf, the number and ratio of BrdU positive cells were obviously reduced in the digestive organs of the sec13sq198 mutant (mu) compared with the wild-type (WT). (A) Transverse section of the wild-type control through the liver (lv), intestine (in) and pancreas (pa). (B) Transverse section of a mutant embryo through the liver (lv) and intestine (in). (C) Transverse section of a mutant through the intestine (in) and pancreas (pa). (D) Histogram showing the ratios of BrdU positive cells versus total cells in a specific organ. Data were obtained from sections derived from three wild-type and three sec13sq198 mutant embryos, respectively. S: somite, L: liver, I: intestine, P: pancreas; **: P<0.01. (E?G) TUNEL assay showing cell apoptosis (cells in red) in wild-type (WT) (E) and mutant (mu) (F) cryosections. Data shown in (G) were obtained by counting apoptotic cells against total cells in a specific organ (e.g., intestine) in sections from three wild-type and three mutant embryos, respectively. *: P<0.05; **: P<0.01.DAPI was to stain the nuclei. G, gut; L, liver; P, pancreas; S, somite. PHENOTYPE:
|
|
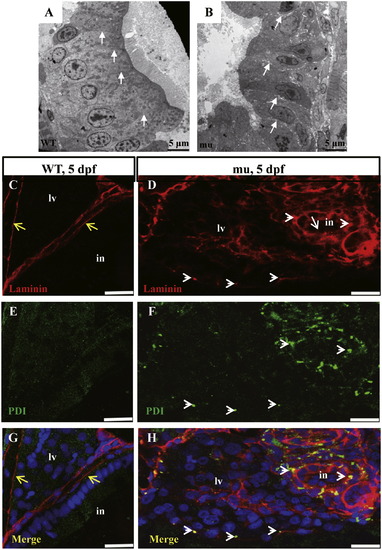
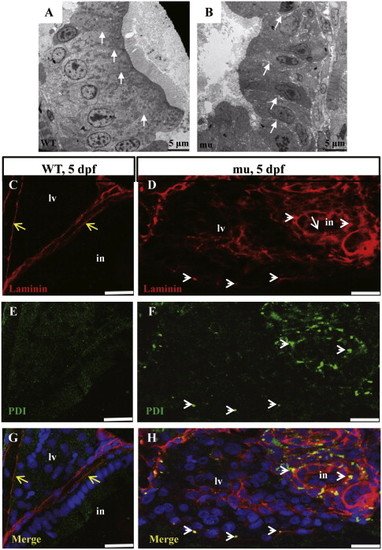
The sq198 mutation causes loosening of the cell-cell junction in the digestive organs. (A,B) TEM analysis of intestinal epithelium in 5 dpf wildtype and mutant embryos. The epithelial cells are tightly adhered to each other in the wildtype (WT) (A, white arrows) but are separated by clearly visible gaps in the mutant (mu) (B, white arrows). (C?H) Double immunofluorescence staining using the antibodies against the basement component Laminin (red) and the ER marker PDI (green). In the wild type control at 5 dpf, Laminin forms the normal lining along the basement layer (C) that nicely separates the liver and intestine (yellow arrows) (G). No apparent accumulation of Laminin in the ER (E,G). In contrast, the mutant did not form an apparent basement layer to separate the liver and intestine (white arrowheads) (D,H) and the mutant cells accumulated Laminin in the ER (white arrowheads) (D,F,H). PHENOTYPE:
|
|
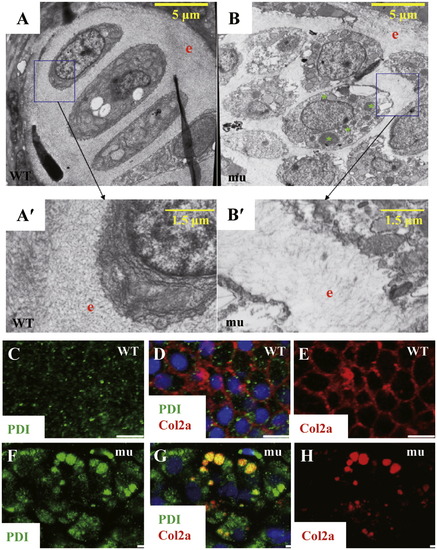
The sq198 mutation impairs the protein secretory pathway. (A,A2,B,B2) TEM analysis. Compared to wildtype (WT), the mutant (mu) exhibited disorganized chondrocytes (compare A and B) and less-accumulated ECM (compare A′ and B′). A′ and B′ represent high resolution pictures of boxed regions in A and B, respectively. e: ECM; green asterisk: unknown vesicles in a mutant chondrocyte. (C?H) Double immunostaning of the ER resident marker PDI and the secretory protein Collagen II (Col2a) in chondrocytes in 5 dpf wildtype (WT) (C?E) and mutant (mu) (F?H) embryos. (C,F) Immunostaining of PDI (green). (E,H) Immunostaining of Col2a secretion (red). (D,G) Superimpose of PDI and Col2a staining accordingly. PHENOTYPE:
|
|
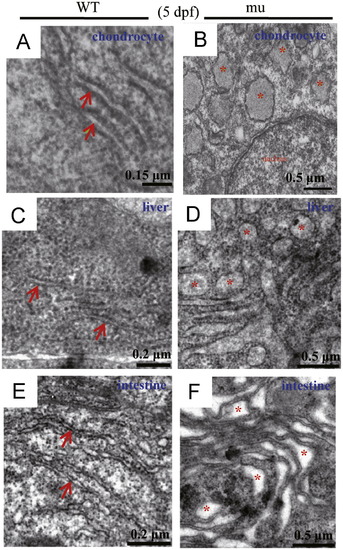
The sq198 mutation disrupts the ER structure in chondrocytes, hepatocytes and intestinal epithelium. (A?F) TEM analysis of the ER structure in chondrocytes (A,B), hepatocytes (C,D), and intestinal epithelium (E,F) in wildtype (WT) and mutant (mu) embryos at 5 dpf. Rough ER in WT cells are indicated with red arrows (A,C,E), disrupted ER structures in the mutant cells are highlighted with red asterisks (B,D,F). PHENOTYPE:
|
|
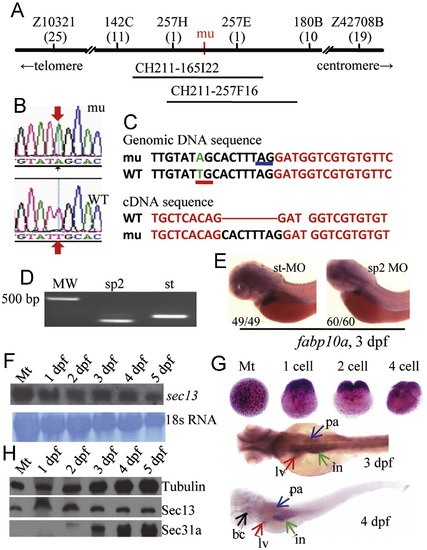
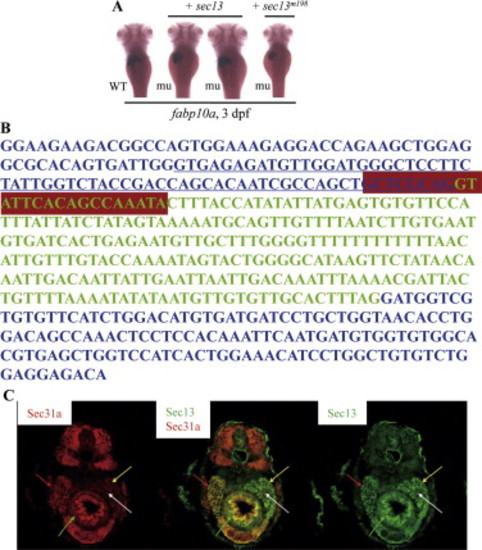
The sec13sq198 mutation creates a novel splice receptor site in intron7 of sec13. (A) Diagram summarizing map-based cloning of the sec13sq198 mutant gene. Genetic markers used in mapping are shown on top with their corresponding number of recombinants shown below in bracket. The mutant gene (mu) was located between markers 257H and 257E spanning a 54.3 kb genomic DNA fragment contained in two overlapping BACs (CH211-165I22 and CH211-257F16). (B,C) Analysis of sec13 genomic DNA sequence in sq198 identified a T to A substitution (indicated by red arrows in B) that creates a novel splice receptor site (underlined in red in C, panel for Genomic DNA sequence) in intron 7 of sec13. Native splice receptor site for Intron7/Exon8: underlined in blue. Intron sequence: black letters; exon sequence: red letters. An eight nucleotides insertion (black letters) was identified in the sec13 cDNA derived from sec13sq198 (C, panel for cDNA sequence). (D,E) Injection of the sec13 splicing blocking morpholino sp2 altered sec13 transcripts with 66 nucleotides deletion (D) that led to the small liver phenotype as conferred by the sec13m198 mutation when examined with the liver marker fabp10a at 3 dpf (E). (F) Northern analysis of sec13 transcripts in unfertilized eggs (Mt) and 1dpf to 5 dpf embryos using a sec13 probe. 18S RNA was used as loading control. (G) WISH analysis of sec13 expression patterns in unfertilized eggs (Mt), zygotic eggs at 1-cell, 2-cell and 4-cell stages, and in embryos at 3 dpf and 4 dpf. bc: branchial arch; in: intestine; lv: liver; pa: pancreas. (H) Western analysis of Sec13 and Sec31a protein expression, respectively, in unfertilized eggs (Mt) and 1 dpf to 5 dpf embryos. Tubulin was used as loading control. PHENOTYPE:
|
|
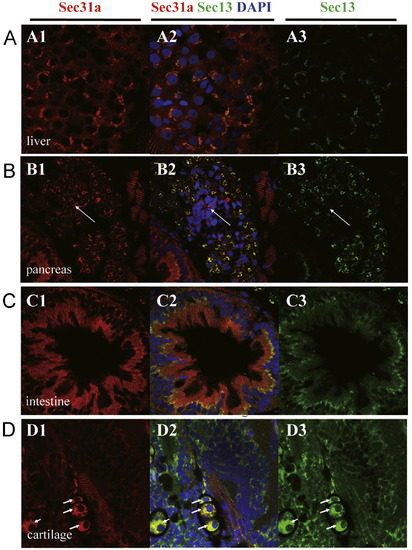
Sec13 and Sec31a co-localize in hepatocytes, exocrine pancreatic cells, intestinal epithelium and chondrocytes. (A?D) In 5 dpf embryos Sec31a and Sec13 co-localize in hepatocytes (A), exocrine pancreatic cells (but not in endocrine pancreatic cells, white arrow) (B), apical side of the intestinal epithelium (C), and chondrocytes (white arrow) (D). (A1,B1,C1,D1) Immunostaining of Sec31a (red). (A3,B3,C3,D3) Immunostaining of Sec13 (green). (A2,B2,C2,D2) Superimpose of Sec31a and Sec13 staining accordingly. DAPI was used to stain the nucleus. EXPRESSION / LABELING:
|
|
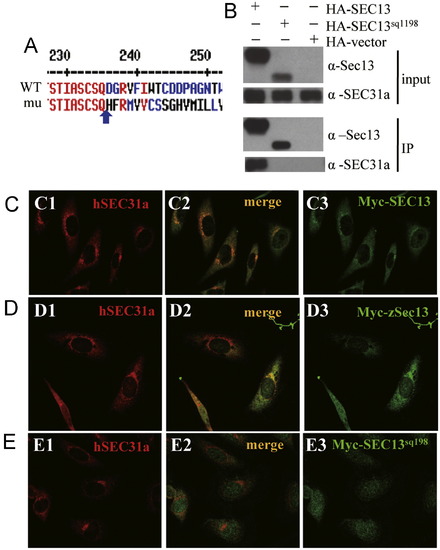
Sec13sq198 mutant protein is a C-terminal truncated Sec13 which is unable to interact with Sec31a. (A) Alignment of the C-terminal sequences of Sec13 and Sec13sq198. Sec13sq198 mutant protein lacks the C-terminus of Sec13 starting from D236 (blue arrow) and contains 34 new amino acids after Q235 (blue arrow). (B) Co-immunoprecipitation (Co-IP) analysis. HEK-293T cells were transfected with HA-SEC13 or HA-SEC13sq198 plasmids. An anti-HA antibody was used to immunoprecipitate (IP) the tagged proteins. The input (1st and 2nd panels) and the IP products (3rd and 4th panels) were blotted using an anti-SEC31A antibody (2nd and 4th panels) and an anti-Sec13 mosue serum (1st and 3rd panels). (C-E) In human Hela cells, Myc-SEC13 (C) or zebrafish Myc-Sec13 (D) co-localizes with endogenous SEC31a whilst SEC13sq198 is evenly distributed in the cytoplasm (E). (C1,D1,E1) Immunofluorescence staining of endogenous SEC31a (red). (C3,D3,E3) Immunostaining of Myc-tagged hSEC13 (C3) or zSec13 (D3) or hSEC13sq198 (E3) (green). (C2,D2,E2) Superimposed. |
|
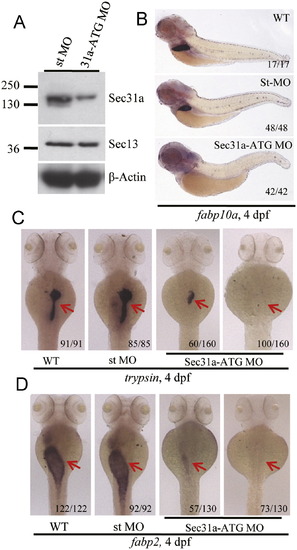
Knockdown of Sec31a resulted in hypoplastic digestive organs. (A) Western blotting analysis of Sec31a and Sec13 in sec31a-ATG MO (31a-ATG MO) and standard control MO (st MO) injected embryos at 4 dpf, respectively. β-Actin was used as the loading control. (B-D) WISH using the fabp10a (B), trypsin (C) and fabp2 (D) probes to examine the development of the liver, exocrine pancreas and intestine in wildtype (WT), standard control MO (st MO), and 31a-ATG MO embryos at 4 dpf. Number of embryos displaying the showed phenotype of the total embryos examined was provided at the bottom right side of each representative embryo. PHENOTYPE:
|
|
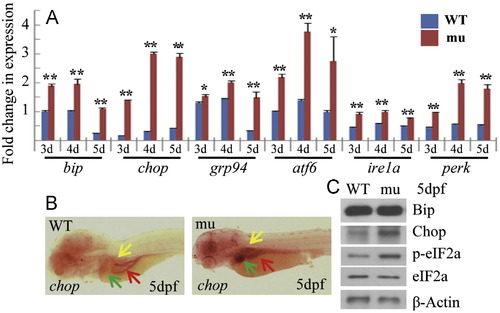
Activation of the UPR pathway in the sec13sq198 mutant. (A) Quantitative RT-PCR analysis of UPR-related genes bip, chop, grp94, atf6, ire1a and perk in WT and the mutant, respectively. The relative expression level of the genes was shown in fold change as normalized to the housekeeping gene, elf1a. (B) WISH analysis of chop expression in WT and mutant (mu) embryos at 5 dpf. (C) Western blot analysis of Bip, Chop, phosphorylated eIF2a (p-eIF2a) and total eIF2a (eIF2a) in WT amd mutant (mu). β-Actin was used as the loading control. EXPRESSION / LABELING:
PHENOTYPE:
|
|
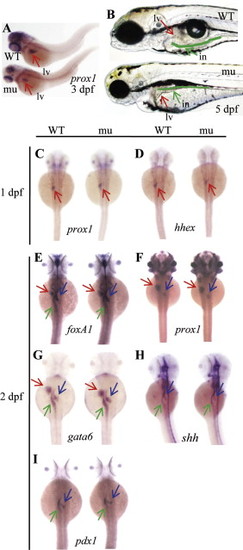
The sq198 mutation affects organ growth but does not affect the specification and early development of major digestive organs. (A) sq198 was identified in our screening as a liver defective mutant using prox1 as a liver marker. (B) sq198 conferred smaller liver, unexpanded swimbladder and much thinner intestine with an unabsorbed yolk as seen under a dissection microscope at 5 dpf. The liver is indicated by a red arrow. The intestinal tract is traced with green line and is indicated with a green arrow. WT: wildtype; mu: mutant; in: intestine; lv: liver. (C-I) WISH using early liver markers prox1 (C,F) and hhex (D), pan-endoderm markers foxA1 (E), gata6 (G) and shh (H), and early pancreatic marker pdx1 (I) on embryos at 1 dpf or 2 dpf as indicated. No observable phenotypic differences between the wildtype (WT) and mutant (mu) for these markers are found during the stages examined. Red arrow: liver; blue arrow: pancreas; green arrow: intestine. |
|
The sec13sq198 mutant embryos develop a small liver but a relatively normal sized islet. WISH using the liver specific marker fabp10a and endocrine pancreas specific marker insulin two probes together showed that although the mutant liver (mu) was marked reduced in its size there was no obvious difference in the sizes of the islets between siblings (sibling) and sec13sq198 mutant (mu) at 3.5dpf.lv: liver; is: islet. PHENOTYPE:
|
|
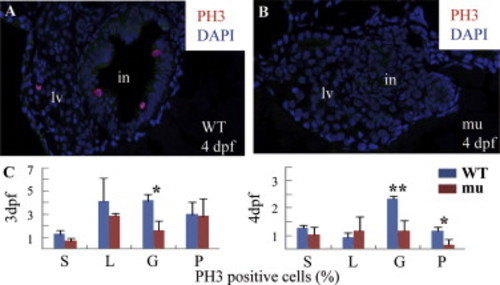
The sq198 mutation causes cell cycle arrest in the digestive organs. (A,B) Histochemical staining using anti-phosphorylated histone 3 (anti-pH3) showing cells entering G2 to M phase in wildtype (WT) (A) and mutant (mu) (B) embryos (cells in red). Pan-cadherin was stained to mark the gut epithelia using a monoclonal antibody against Pan-cadherin (Sigma, C1821) in dilution of 1:200 (green). (C) The ratios of pH3-positive cells shown in different tissues were obtained by counting pH3-positive cells against total cells in a specific organ (e.g., intestine) in sections obtained from three wild-type and three mutant embryos, respectively. *: P<0.05; **: P<0.01. PHENOTYPE:
|
|
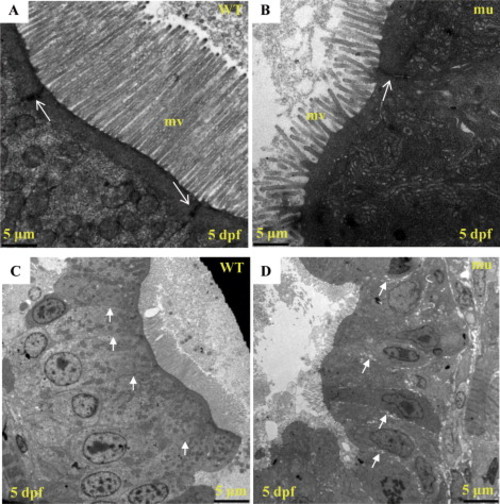
The sq198 mutation disorganizes intestinal epithelium. (A,B) TEM analysis of intestinal epithelium in wildtype (WT) and mutant (mu) embryos at 5 dpf. Mutant exhibits shortened and less organized microvilli (mv). Meanwhile, electron dense material is greatly reduced and narrowed in the apical cell-cell junction area in the mutant (white arrow). (C,D) An enlarged view of Fig. 2G and 2H, respectively. PHENOTYPE:
|
|
Rescue and mimicking of sec13m198 mutant phenotype. (A) Injection of the wildtype sec13 mRNA (+ sec13) but not the mutant mRNA (+ sec13m198) could fully or partially rescue the sec13m198 small liver phenotype when examined with an fabp10a probe on embryos 3 days post-injection. WT: wildtype; mu: mutant. (B) Injection of the sec13 splicing blocking morpholino sp2 resulted in a transcript with 66 nucleotides deletion originated from exon 7 (underlined sequence). Total RNA for RT-PCR was extracted from embryos at 3 dpf.RT-PCR product was then sequenced. Exons 7 and 8 of sec13 are in blue letters while intron 7 is in green. sp2 targeting sequence is boxed in purple. st: standard control morpholino; MO: sp2 morphant; MW: molecular weight. (C) Sec13 and Sec31a are co-localized in digestive organs. Immunofluorescence staining analysis using an anti-Sec13 mouse serum (panel on the right, green) and an anti-SEC31a rabbit serum (panel on the left, red) showed that Sec13 and Sec31A expression are co-localized in major digestive organs in 5 dpf embryos (panel in the middle). Red arrow: liver; yellow arrow: exocrine pancreas; white arrow: endocrine pancreas; green arrow: intestine. EXPRESSION / LABELING:
|
|
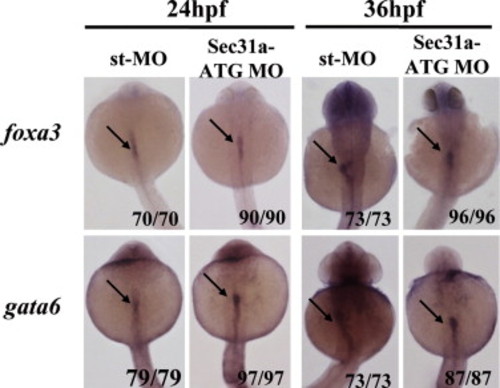
Knock-down of Sec31a-ATGMO did not block the endoderm initiation. WISH using early endoderm markers foxa3 and gata6 on embryos at 24 and 36 hpf showed that the endoderm rod was formed in the Sec31a-ATGMO morphants. Arrow points to the endoderm rod. |
|
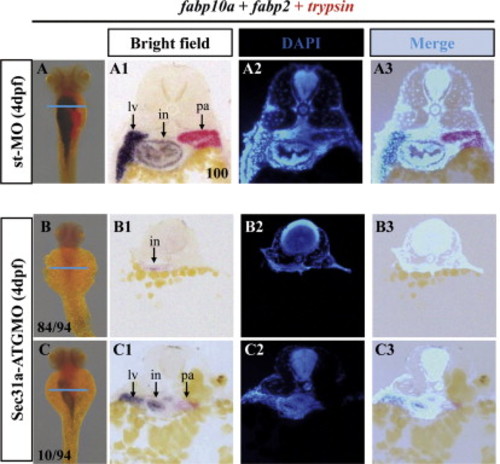
Majority of Sec31a-ATGMO exhibited greatly diminished but not abolished endoderm-derived tissues. (A-C) WISH using Dig-labeled fabp10a and fabp2 (dark blue) and Fast Red-labeled trypsin (red) probes showed that, when compared with that in the control (st-MO) at 4dpf (A), Sec31a-ATGMO morphants were compromised in the development of the digestive organs, with majority (84/94) displayed dramatically diminished organs (B) and the rest (10/94) small organs (C). Cross sectioning of the embryos after WISH (along the blue line as indicated, A1-A3 sections: corresponding to A; B1-B3 sections: to B; C1-C3 sections: to C) showed that the fabp2 positive cells were greatly reduced in number but were not abolished in the type of Sec31a-ATGMO morphants (B). PHENOTYPE:
|
|
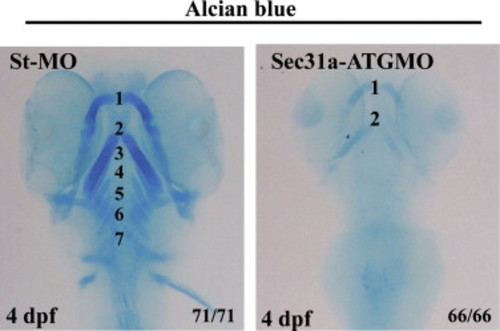
Sec31a-ATGMO morphants exhibited malformed branchial cartilage. Alcian blue staining showed that, compared with the branchial arches 1-7 in the control embryos (st-MO), the structure of the branchial cartilage is disrupted in Sec31a-ATGMO morphants at 4 dpf. Number of embryos examined is shown at the right corner of the bottom. PHENOTYPE:
|
|
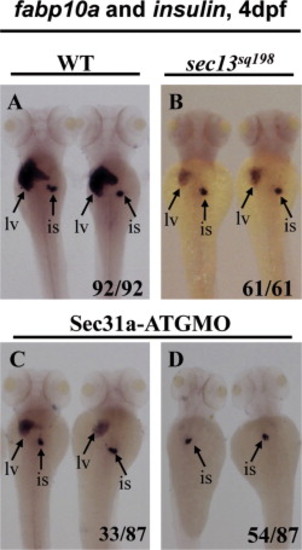
Both Sec13 and Sec31a are dispensable for the islet development. (A-D) WISH using the liver specific marker fabp10a and endocrine pancreas specific marker insulin as probes showed that all sec13sq198 mutant embryos and Sec31a-ATGMO morphants developed an islet that is not obvious different from that in the wild type control at 4 dpf. However, compared to the wild-type control (WT) (A), all sec13sq198 mutant embryos (mu) (61/61) (B) and Sec31a-ATGMO morphants (33/87) examined (C) developed a small liver while majority of Sec31a-ATGMO morphants (54/87) appeared to be liverless (D). lv: liver; is: islet. PHENOTYPE:
|
|
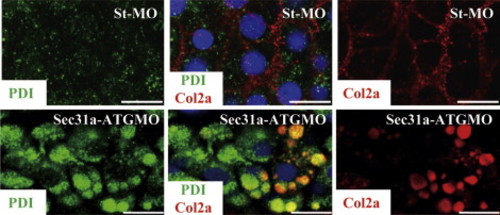
Knockdown of Sec31a disrupted ER structure. Double immunostaning of the ER resident marker PDI (Sigma, P7496-200ul, in 1:200 dilution) (green) and Col2a (Lifespan, LS-C41831, in 1:100 dilution) (red) in chondrocytes in 4.5 dpf St-MO morphant (top panels) and Sec31a-ATGMO morphant (bottom panels). PHENOTYPE:
|
|
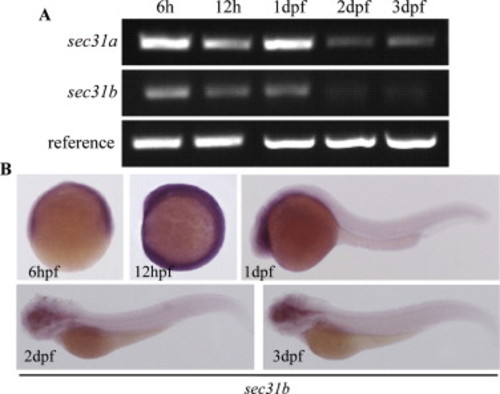
Analysis of sec31b expression patterns. (A) Semi-quantitative RT-PCR analysis of sec31b and sec31b transcripts at the stages of 6 hpf, 12 hpf, 1 dpf, 2 dpf and 3 dpf. First strand cDNA (reverse transcription, RT) was synthesized from total RNA by MMLV reverse transcriptase (Invitrogen) using oligo-dT primers and then used as the template in PCR. PCR was run at 28 cycles of 94oC (30 second), 55oC (30 second), 72oC (20 second). For analyzing sec31a: forward primer: AgCCgTgTCTTCTggAgggTgTT, reverse primer: ggTgCgggAggAgTgAAgTgTTg, For analyzing sec31b: forward primer: ATgAAgCgCgTgTgAAgTTTTTgA, reverse primer: TACggggATCTggAAgggCTgTTT, (B) WISH analysis of sec31b expression at the stages of 6 hpf, 12 hpf, 1 dpf, 2 dpf and 3 dpf. RT product from 1 dpf was used as the template to clone sec31b using primer pair ACgTgAATTCCTCCgCCCCAgCAACAACAACC (forward) and ACgTTCTAgACCCCgAgCAgATgAAgCACAgACA (reverse). PCR product was digested with EcoRI and Xbal1 and then cloned to the pCS2+ vector for generating DIG-labeled WISH riboprobe. EXPRESSION / LABELING:
|
Reprinted from Developmental Biology, 367(2), Niu, X., Gao, C., Jan Lo, L., Luo, Y., Meng, C., Hong, J., Hong, W., and Peng, J., Sec13 safeguards the integrity of the endoplasmic reticulum and organogenesis of the digestive system in zebrafish, 197-207, Copyright (2012) with permission from Elsevier. Full text @ Dev. Biol.