- Title
-
Zebrafish smarcc1a mutants reveal requirements for BAF chromatin remodeling complexes in distinguishing the atrioventricular canal from the cardiac chambers
- Authors
- Auman, H.J., Fernandes, I.H., Berríos-Otero, C.A., Colombo, S., Yelon, D.
- Source
- Full text @ Dev. Dyn.
|
The sk29 mutation causes multiple embryonic defects. (A, B) Lateral views of live embryos at 48 hpf, anterior to the left, dorsal to the top. In comparison to wild-type embryos (wt), sk29 mutant embryos (mut) have a slightly shorter body axis, decreased pigmentation, pericardial edema, and a thickened yolk extension. (C, D) Lateral views at 48 hpf, anterior to the top, dorsal to the left. In sk29 mutants, cardiac chambers (arrowheads) fail to expand and remain relatively linear, with little apparent morphological distinction of the AVC. (E, F) Dorsal views at 72 hpf, anterior to the top. Pectoral fins (arrows) are absent (asterisks) in sk29 mutants. (G, H) Lateral views at 48 hpf, anterior to the top, dorsal to the left. Retinal pigmentation is abnormal in sk29 mutants. PHENOTYPE:
|
|
The sk29 locus encodes smarcc1a. (A) Schematic depicting meiotic map of the sk29 critical interval. The sk29 mutation is flanked by SSLP markers z10632 and z6293 on chromosome 16 and is tightly linked to SSLP marker z1215 (no recombinants out of 2350 meioses). Additional SNP and SSLP markers (ensembl.org/Danio_rerio) near the sk29 mutation are indicated with numbers of recombinants (r) out of 2350 meioses. (B) cDNA from sk29 mutants contains a C to T transition at position 1942 of the smarcc1a open reading frame (arrows), resulting in a premature stop codon. (C) Zebrafish smarcc1a encodes a 1089 amino acid protein with several conserved motifs and approximately 74% overall similarity to human SMARCC1. The SMART domain classification program63 identifies Chromo64 and SANT26 domains within residues 211-258 and 613-661, respectively. Pfam classification65 identifies a SWIRM domain66 within residues 444-532. The smarcc1ask29 nonsense mutation changes Gln648 (CAA) to a stop codon (TAA), disrupting the SANT domain. (D?G) The sk29 mutation causes a strong reduction in levels of smarcc1a transcripts. Lateral views, anterior to the top. (D, F) Whole-mount in situ hybridization indicates that the smarcc1a expression pattern is not spatially restricted in wild-type. (E, G) smarcc1a transcripts are dramatically reduced in smarcc1ask29 mutants at the 10 and 21 somite (som) stages. (H?K) The sk29 mutation causes a strong reduction in levels of Smarcc1 protein. (H, I) Lateral views, anterior to the right, dorsal to the top. (J, K) Lateral views, anterior to the left, dorsal to the top. Whole-mount immunostaining shows presence of Smarcc1 protein throughout the wild-type embryo at 16 som (H) and within the wild-type heart at 36 hpf (J, arrowhead). Smarcc1 immunostaining is dramatically reduced in sk29 mutants at 16 som (I) and 36 hpf (K). EXPRESSION / LABELING:
PHENOTYPE:
|
|
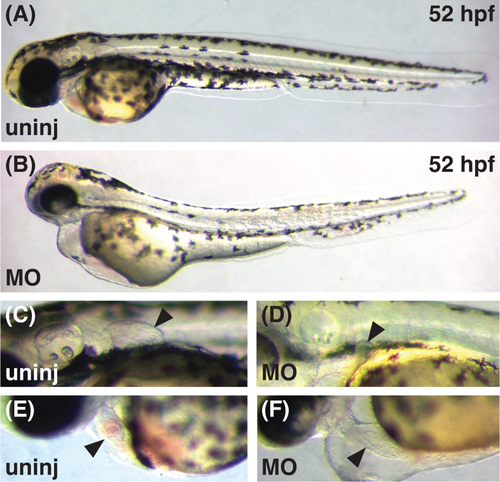
Morpholino knockdown of smarcc1a phenocopies smarcc1ask29 mutants. (A, B) Lateral views of live embryos at 52 hpf, anterior to the left, dorsal to the top. Morpholino-injected embryos (MO) exhibit reduced pigmentation, pericardial edema, and abnormal retinal development. (C, D) Pectoral fins (arrowheads) do not develop normally in MO-injected embryos, resulting in small, stunted fin buds at 52 hpf. (E, F) In contrast to the heart of an uninjected wild-type embryo (arrowhead, E), the heart of a MO-injected embryo (arrowhead, F) has dysmorphic chambers aligned in a linear arrangement. Overall, the phenotypes of MO-injected embryos are highly similar, although somewhat less severe, than those of smarcc1ask29 mutant embryos, likely reflecting the limited efficiency of morpholino knockdown. PHENOTYPE:
|
|
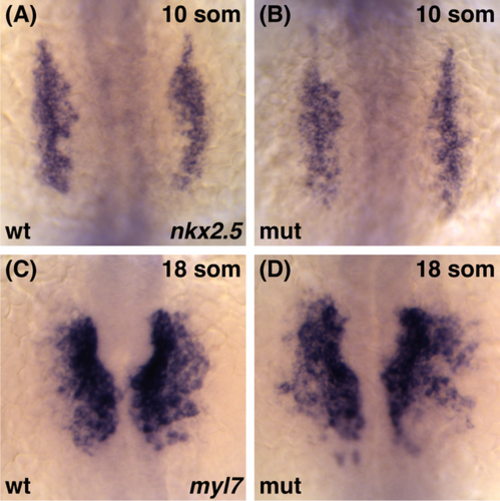
Cardiomyocytes differentiate normally in smarcc1a mutants. Whole-mount in situ hybridization depicts the expression of nkx2.5 (A, B) and myl7 (C, D) in the ALPM at 10 and 18 som, respectively. Dorsal views, anterior to the top. (A, B) The nkx2.5 expression domain in smarcc1a mutant embryos is indistinguishable from that of wild-type embryos. (C, D) smarcc1a mutants do not exhibit evident deficiencies in their populations of myl7-expressing differentiated cardiomyocytes. However, the morphogenesis of the medially migrating cardiomyocytes appears slightly delayed and disorganized in smarcc1a mutants EXPRESSION / LABELING:
PHENOTYPE:
|
|
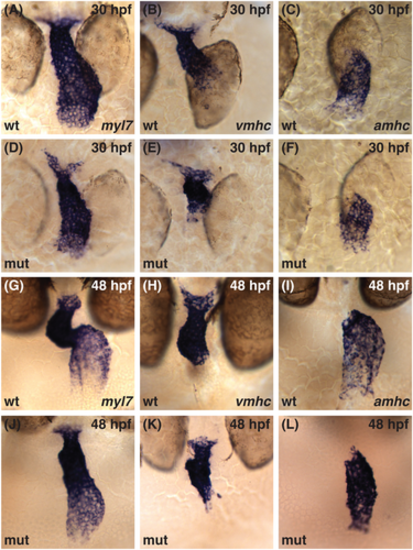
Cardiac chamber identity is established normally in the smarcc1a mutant heart tube. (A?L) Whole-mount in situ hybridization compares expression of myl7 in all cardiomyocytes (A, D, G, J) with expression of vmhc in ventricular cardiomyocytes (B, E, H, K) and amhc in atrial cardiomyocytes (C, F, I, L). (A?F) Dorsal views at 30 hpf, ventricle to the top. (G?L) Frontal views at 48 hpf, ventricle to the top. vmhc and amhc expression are found in complementary domains corresponding to ventricular and atrial identities in both wild-type embryos and smarcc1a mutants within the linear heart tube (A?F) and as development proceeds (G?L). |
|
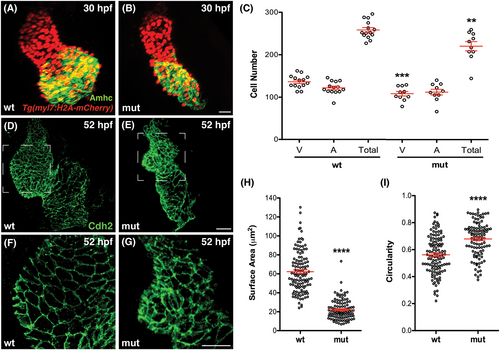
Number and shape of cardiomyocytes are altered in smarcc1a mutants. (A, B) Immunofluorescence for Amhc (green) and mCherry (red) in wild-type (A) and smarcc1a mutant (B) hearts expressing Tg(myl7:H2A-mCherry) at 30 hpf. (C) Graphs depict the number of cardiomyocytes in the ventricle (V) and atrium (A), as well as the total number of cardiomyocytes, in wild-type and smarcc1a mutant hearts at 30 hpf. Mean and SE of each data set are shown. Asterisks indicate statistically significant differences from wild-type, using an unpaired t-test (**P < .01, ***P < .001). Number of hearts analyzed: wt, n = 14; smarcc1a, n = 10. The smarcc1a mutant heart contains slightly fewer ventricular cells than the wild-type heart at 30 hpf. (D?G) Cdh2 immmunostaining (green) outlines cardiomyocytes in wild-type (D, F) and smarcc1a mutant (E, G) hearts at 52 hpf. (F, G) Enlarged views of the ventricles shown in (D, E). Wild-type chamber emergence is accompanied by the increased surface area and elongation of cardiomyocytes along the outer curvature of the ventricle. In smarcc1a mutants, ventricular outer curvature cardiomyocytes do not enlarge or elongate normally and instead retain relatively small and round cell surfaces. (H, I) Graphs depict surface area (H) and circularity (I) of individual cardiomyocytes in the outer curvature of wild-type and smarcc1a mutant ventricles at 52 hpf. Mean and SE of each data set are shown. Asterisks indicate statistically significant differences from wild-type, using an unpaired t-test (****P < .0001). Number of hearts analyzed: wt, n = 3; smarcc1a, n = 3. Ventricular outer curvature cardiomyocytes in smarcc1a mutants are smaller (H) and rounder (I) than their wild-type equivalents at 52 hpf. |
|
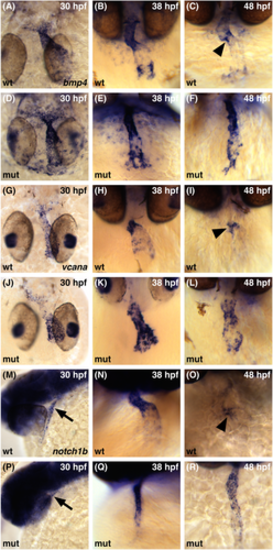
smarcc1a is required to restrict patterns of gene expression to the AVC. Whole-mount in situ hybridization compares expression patterns of bmp4 (A?F), vcana (G?L), and notch1b (M?R) expression between 30 and 48 hpf. (A, D, G, J) Dorsal views, ventricle to the top. (B, C, E, F, H, I, K, L, N, O, Q, R) Frontal views, ventricle to the top. (M, P) Lateral views, anterior to the left; arrows indicate the ventricular end of the heart tube. (A, D, G, J, M, P) The initial broad expression of all three genes is similar in wild-type and smarcc1a mutant embryos. (B, E) In smarcc1a mutants at 38 hpf, the expression of bmp4 continues to resemble wild-type expression, with higher levels in the ventricle and inflow tract and lower levels in the atrium. (H, K) Expression of vcana is perturbed in smarcc1a mutants at 38 hpf, with high levels found throughout the heart. (N, Q) The expression of notch1b in smarcc1a mutants at 38 hpf is higher in the ventricular endocardium than in the atrial endocardium, similar to the pattern seen in wild-type. (C, F, I, L, O, R) In wild-type at 48 hpf, the refined patterns of bmp4, vcana, and notch1b feature concentrated expression in the AVC (arrowheads in C, I, O), and this refinement does not occur in smarcc1a mutants. |
|
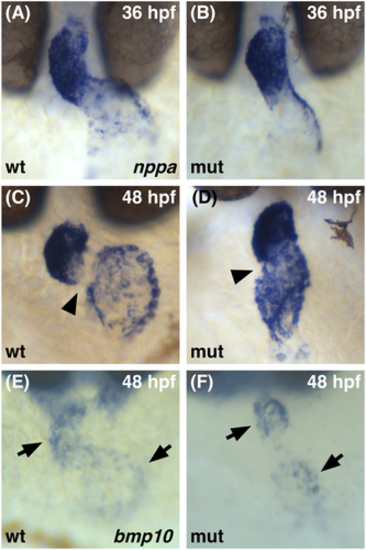
Normal initiation of gene expression in the chamber myocardium of smarcc1a mutants. Whole-mount in situ hybridization compares expression patterns of nppa (A?D) and bmp10 (E, F) expression at 36 (A, B) or 48 (C?F) hpf. Dorsal views, ventricle to the top. (A, B) The initial expression of nppa is similar in wild-type and smarcc1a mutant hearts at 36 hpf. (C, D) At 48 hpf, wild-type expression of nppa is found in the chamber myocardium but not in the AVC (arrowhead). In smarcc1a mutants, elevated expression is seen throughout the heart, including in the AVC (arrowhead). (E, F) In both wild-type and smarcc1a mutant hearts at 48 hpf, bmp10 expression is found in the cardiac chambers (arrow), with a clear gap in expression at the AVC. |
|
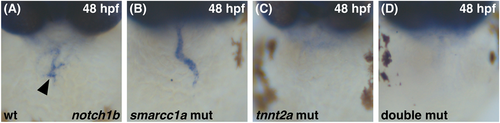
Comparison of smarcc1a mutant and tnnt2a mutant phenotypes. Whole-mount in situ hybridization compares expression patterns of notch1b in sibling embryos at 48 hpf. Dorsal views, ventricle to the top. (A) In wild-type, notch1b expression is concentrated in the AVC endocardium (arrowhead). (B) In smarcc1a mutants, notch1b expression is found throughout the endocardium. (C) In tnnt2a mutants, notch1b expression is not detectable in the endocardium. (D) In smarcc1a;tnnt2a double mutants, notch1b expression is also not detectable in the endocardium. |
|
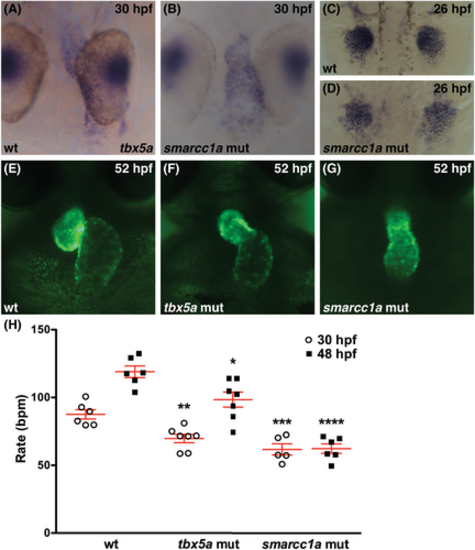
Loss of tbx5a function is not sufficient to account for the smarcc1a cardiac phenotype. (A?D) Whole-mount in situ hybridization depicts expression of tbx5a in wild-type or smarcc1a mutant embryos. (A, B) Dorsal views at 30 hpf, ventricle to the top. (C, D) Dorsal views at 26 hpf, anterior to the top. Expression of tbx5a appears normal in the heart tube (A, B) and pectoral fin bud mesenchyme (C, D) of smarcc1a mutants, although coalescence and growth of the fin mesenchyme is altered. (E?G) Frontal views of live embryos expressing Tg(myl7:egfp)at 52 hpf. Cardiac chamber shape and looping are more severely affected in smarcc1a mutants (G) than in tbx5a mutants (F). (H) Graph depicts heart rate (beats per minute, bpm) in wild-type, tbx5a mutant, and smarcc1a mutant embryos at 30 and 48 hpf. Mean and SE of each data set are shown. Asterisks in graph indicate statistically significant differences between tbx5a mutant and wild-type heart rates or between smarcc1a mutant and wild-type heart rates at each time point, using an unpaired t-test (*P < .05; **P < .01; ***P < .001; ****P < .0001). Number of hearts analyzed at 30 hpf: wt, n = 6; tbx5a, n = 7; smarcc1a, n = 5. Number of hearts analyzed at 48 hpf: wt, n = 6; tbx5a, n = 7; smarcc1a, n = 6. Heart rate in smarcc1a mutants is significantly lower than in tbx5a mutants at 48 hpf (P < .001, t-test), and the smarcc1a mutant heart rate does not increase between 30 and 48 hpf. |