- Title
-
Zebrafish Cdx1b modulates epithalamic asymmetry by regulating ndr2 and lft1 expression
- Authors
- Wu, C.S., Lu, Y.F., Liu, Y.H., Huang, C.J., Hwang, S.L.
- Source
- Full text @ Dev. Biol.
|
Fig. 1. cdx1b expression during gastrulation and segmentation stages. RNA in situ hybridization was conducted on embryos from 2-cell (A), sphere (B), shield (C), and bud (D, E) stages using a cdx1b antisense RNA probe. Enriched cdx1b expression in the shield (C, arrow) and forebrain neural keel (E) is shown. Double in situ hybridization was performed on embryos from 2 ?s (F) stage using cdx1b (blue) and lft1 (red) RNA probes as well as 24 ?s (H, I) stage using cdx1b (red) and lft1 (green) RNA probes. A sagittal section of stained embryos (G) is shown. Double in situ hybridization was conducted on 22 ?s embryos with cdx1b (blue) and flh (red) probes (J) or with cdx1b (red) and otx5 (green) RNA probes (K). Scale bar (G) represents 100 ??m; scale bar (H, I) represent 40 ??m ?dd, diencephalon; fnk, forebrain neural keel; h, heart; pcp, prechordal plate. EXPRESSION / LABELING:
|
|
Fig. 2. A small but significant portion of Zcdx1b?/? mutant embryos exhibited absent pitx2c expression in the left dorsal diencephalon. A The nucleotide sequences and protein domains of wild-type and Zcdx1b?/? mutants are shown. Deleted nucleotides are indicated by a dashed line, while inserted nucleotides are shown in red lettering. The sgRNA target site in the activation domain is highlighted in yellow and the protospacer adjacent motif (PAM) sequence is shown in blue. B Nucleotide sequences of wild-type (WT) and Zcdx1b?/? mutants (MT) within the sgRNA target region are shown. C Percentages of embryos with left, right, bilateral or absent expression patterns of pitx2c in the dorsal diencephalon from intercross of Zcdx1b+/- F2 mutants are shown. Data were combined from two different intercrossing experiments. Chi-squared test of independence was conducted to compare portions of embryos with absent pitx2c expression in the dorsal diencephalon between sibling wild-type and Zcdx1b+/- or Zcdx1b?/? mutant embryos. ?, p ?< ?0.05. |
|
Fig. 3. The majority of MZcdx1b?/? mutant embryos showed absent expression of ndr2, lft1 and pitx2c in the left dorsal diencephalon and absent expression of lft1 in the heart at segmentation and pharyngula stages. RNA in situ hybridization was conducted on wild-type (WT) or MZcdx1b?/? mutant embryos to evaluate expression patterns in the dorsal diencephalon or heart using ndr2 (A), lft1 (B, C), lft2 (D) or pitx2c (E) RNA probes at 19?22 ?s or 26 hpf stages. Quantitative data show the percentages of embryos displaying different expression patterns of ndr2, lft1 or pitx2c in the dorsal diencephalon or lft1 expression pattern in the heart in wild-type and MZcdx1b?/? mutants. Chi-squared test of independence was conducted to compare proportions of embryos with absent ndr2, lft1 or pitx2c expression patterns in the dorsal diencephalon or absent expression of lft1 in the heart between wild-type and MZcdx1b?/? mutant embryos. ??, p ?< ?0.01; ???, p ?< ?0.001. Scale bar represents 100 ??m ?dd, diencephalon; h, heart, l; left; r, right. EXPRESSION / LABELING:
PHENOTYPE:
|
|
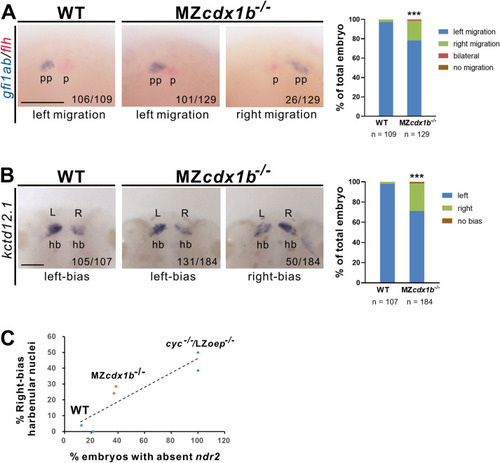
Fig. 4. MZcdx1b?/? mutants showed aberrant parapineal migration and habenular laterality. RNA in situ hybridization was conducted on wild-type (WT) and MZcdx1b?/? mutant embryos using gfi1ab (blue) and flh (red) (A) or kctd12.1 (B) RNA probes at 72 hpf. Quantitative data show the percentages of embryos displaying different positions of parapineal or kctd12.1 expression intensities in the left or right habenula between wild types and MZcdx1b?/? mutants. Scale bar represents 100 ??m. C The correlation between absent ndr2 expression in the dorsal diencephalon and right-biased habenular subnuclei pattern was analyzed for cyc?/?/LZoep?/?, MZcdx1b?/? and wild-type embryos by linear regression. Y ?= ?0.4546X + 0.7175, R2 ?= ?0.8197. Chi-squared test of independence was conducted to compare the proportions of embryos with right-side parapineal position or intense kctd12.1 expression intensity in the right habenula between wild types and MZcdx1b?/? mutants. ???, p ?< ?0.001. hb, habenula; l, left; p, pineal; pp, parapineal; r, right. EXPRESSION / LABELING:
PHENOTYPE:
|
|
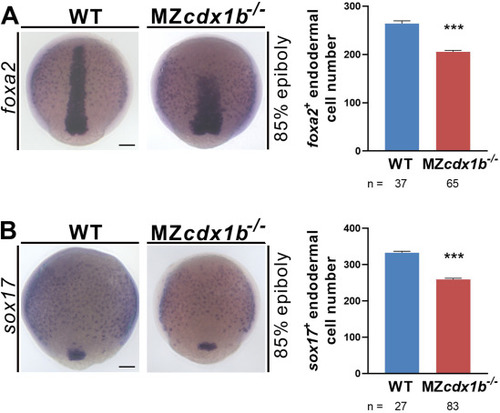
Fig. 5. MZcdx1b?/? mutant exhibited significant reduction in foxa2-and sox17-expressing endodermal precursors. RNA in situ hybridization was conducted on wild-type (WT) and MZcdx1b?/? mutant embryos at 85% epiboly using foxa2 (A) or sox17 (B) RNA probes. Quantification of respective numbers of foxa2-or sox17-expressing endodermal precursors among wild types and MZcdx1b?/? mutants is shown. Scale bar represents 100 ??m. Student?s t-test was conducted. ???, p ?< ?0.001. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Fig. 6. Cdx1b directly binds to CDX binding motifs located upstream of ndr2 and lft1 genes. A Genomic structure and nucleotide sequences of CDX binding element (CBE) (?189 to ?1 bp) and non-binding control region (+1069 to +1256 bp) of the ndr2 gene are shown. One or two CDX binding motifs located in the exon coding region as non-binding control or CBE are highlighted with yellow. B Genomic structure and nucleotide sequences of CBE I (?1825 to ?1634 bp) and CBE II (?2653 to ?2476 bp) of the lft1 gene are shown. Two or three CDX binding motifs located in the CBE I or CBE II are indicated by yellow highlighting. C Chromatin DNA from cdx1b-Myc-injected bud embryos was immunoprecipitated using anti-Myc antibody or anti-IgG antibody as a control. The immunoprecipitated DNA was used for qPCR with specific primers spanning the CBE or non-binding control region of ndr2. The CDX binding motifs within CBE but not non-binding control were significantly enriched by anti-Myc antibody compared with IgG control. D The immunoprecipitated chromatin DNA isolated from cdx1b-Myc-injected bud embryos was subjected to qPCR using specific primers spanning CBE I or CBE II of lft1. The CDX motifs within CBE I but not CBE II were significantly enriched by anti-Myc antibody compared with IgG control. Error bars indicate the standard deviation. Student?s t-test was performed. ?, p ?< ?0.05. |
|
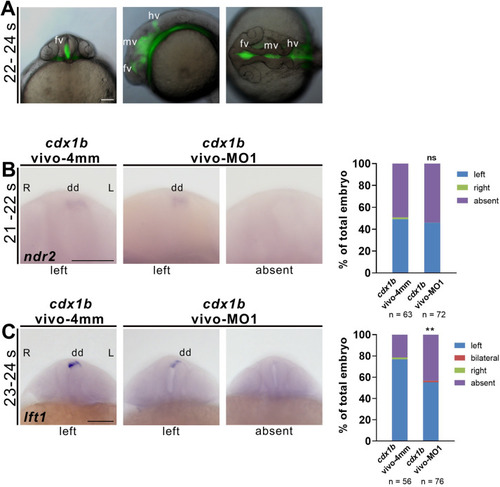
Fig. 7. Cdx1b regulates expression of ndr2 and lft1 in the dorsal diencephalon. A Images of a 22?24 ?S embryo that was injected with cdx1b-vivo-MO1 and dextran-fluorescein in its ventral, lateral and dorsal positions. B RNA in situ hybridization was conducted on cdx1b-vivo-4mm MO1-(cdx1b-vivo-4mm) or cdx1b-vivo-MO1-injected embryos to evaluate expression pattern in the dorsal diencephalon using ndr2 RNA probes at 21?22 ?s stage. C RNA in situ hybridization was conducted on cdx1b-vivo-4mm MO1- or cdx1b-vivo-MO1-injected embryos to evaluate expression pattern in the dorsal diencephalon using lft1 RNA probes at 23?24 ?s stage. Quantitative data show the percentages of embryos displaying different expression patterns of ndr2 or lft1 in the dorsal diencephalon in cdx1b-vivo-4mm MO1- and cdx1b-vivo-MO1-injected embryos. Embryos (B, C) are shown in ventral view. Scale bar represents 100 ??m. Chi-squared test of independence was conducted to compare proportions of embryos with absent ndr2 or lft1 expression patterns in the dorsal diencephalon between cdx1b-vivo-4mm MO1- and cdx1b-vivo-MO1-injected embryos. ??, p ?< ?0.01; ns, not significant. dd, diencephalon; fv, forebrain ventricle; hv, hindbrain ventricle; L, left; mv, midbrain ventricle; R, right. EXPRESSION / LABELING:
PHENOTYPE:
|
|
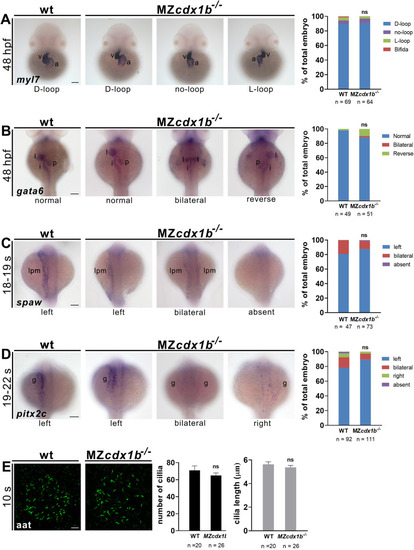
Fig. 8. MZcdx1b?/? mutants display normal visceral organ left-right patterning and left-side expression of spaw and pitx2c as well as normal KV cilia number and length. RNA in situ hybridization was conducted on wild types (WT) and MZcdx1b?/? mutants to evaluate expression patterns in the heart (A) or digestive organs (B) using myl7 (A) or gata6 (B) RNA probes at 48 hpf. Quantitative data show the percentages of embryos displaying different expression patterns of myl7 in the heart or gata6 in various digestive organs of wild types and MZcdx1b?/? mutants. RNA in situ hybridization was conducted on wild types and MZcdx1b?/? mutants to evaluate expression pattern of spaw in the LPM (C) or pitx2c in the gut (D) during 18?19 ?s or 19?22 ?s stages. Quantitative data show the percentages of embryos displaying different expression patterns of spaw in the LPM or pitx2c in the gut in wild types and MZcdx1b?/? mutants. E KV cilia were visualized by immunofluorescence staining using anti-acetylated tubulin (aat) antibody at 10 ?s. Quantitative data show similar numbers and lengths of KV cilia in wild types and MZcdx1b?/? mutants. Embryos (A?D) are shown in dorsal view. Scale bars (A?D) represent 100 ??m; scale bar (E) represents 10 ??m. Chi-squared test of independence was conducted to compare expression patterns of myl7 in the heart, gata6 in the digestive organs, spaw in the LPM or pitx2c in the gut, between wild types and MZcdx1b?/? mutants. Student?s t-test was conducted to compare KV cilia number or length between wild types and MZcdx1b?/? mutants. ns, not significant. a, atrium; g, gut; i, intestine; l, liver; lpm, lateral plate mesoderm; p, pancreas; v, ventricle. EXPRESSION / LABELING:
PHENOTYPE:
|
|
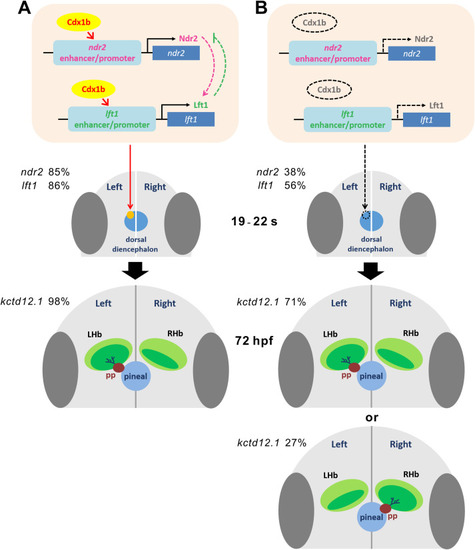
Fig. 9. A proposed model describing the role of Cdx1b in the regulation of epithalamic laterality. A Cdx1b may activate and maintain expression of ndr2 and lft1 in the left dorsal diencephalon during 19?22 ?s, which is essential for subsequent left-sided parapineal migration and asymmetry of habenular nuclei at 72 hpf. Percentages of ndr2 and lft1 expression in the left dorsal diencephalon and left-bias expression of kctd12.1 in the habenula are indicated in wild-type embryos. B In MZcdx1b?/? mutant embryos, reduced and absent expression of ndr2, lft1 and pitx2c in the left dorsal diencephalon results in aberrant parapineal migration and habenular laterality. Percentages of absent expression of ndr2 and lft1 in the dorsal diencephalon as well as left-bias or right-bias expression of kctd12.1 in the habenula are indicated in MZcdx1b?/? mutant embryos. |

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. EXPRESSION / LABELING:
PHENOTYPE:
|
Reprinted from Developmental Biology, 470, Wu, C.S., Lu, Y.F., Liu, Y.H., Huang, C.J., Hwang, S.L., Zebrafish Cdx1b modulates epithalamic asymmetry by regulating ndr2 and lft1 expression, 21-36, Copyright (2020) with permission from Elsevier. Full text @ Dev. Biol.