- Title
-
Dynamic coupling of pattern formation and morphogenesis in the developing vertebrate retina
- Authors
- Picker, A., Cavodeassi, F., Machate, A., Bernauer, S., Hans, S., Abe, G., Kawakami, K., Wilson, S.W., and Brand, M.
- Source
- Full text @ PLoS Biol.
|
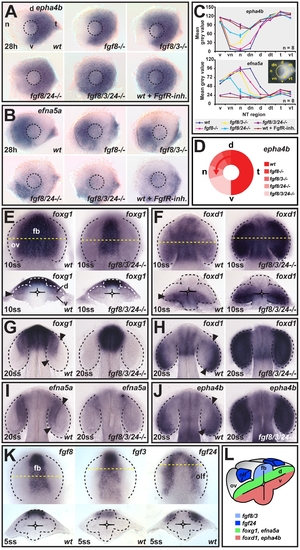
fgf8/3/24 impose nasal-temporal pattern by signaling along the dorsal-ventral axis of the optic vesicle. (A and B) Expression of (A) epha4b in the temporal and (B) efna5a in the nasal retina of wild-type (wt) control, fgf8 mutant (fgf8-/-), fgf8/3 double mutant (fgf8/3-/-), fgf8/24 double mutant (fgf8/24-/-), fgf24-morpholino injected fgf8/3 double mutant (fgf8/3/24-/-), and wt embryos after Fgf receptor inhibitor treatment (wt+FgfR-inh.) at 28 h (nasal is to the left and dorsal is up). Dashed-line circle indicates position of the lens. (C) Nasal-temporal profile of epha4b and efna5a gene expression levels (mean grey value for n = 8 images analyzed for each genotype, y-axis, in defined nasal-temporal (NT) regions of the retina, x-axis, see inset, one representative standard deviation is plotted). (D) Schematic illustrating the graded, nasal expansion of the temporal marker epha4b in the phenotypic series of fgf mutants. (E and F) Expression of the future nasal marker foxg1 (E) and the future temporal marker foxd1 (F) at 10ss in the optic vesicle (ov) of wt (left) and fgf8/3/24-/- embryos (right) (top: dorsal view, anterior to the top, bottom: cross-section, dorsal to the top). (G?J) Expression of the future nasal markers foxg1 (G) and efna5a (I) and the future temporal markers foxd1 (H) and epha4b (J) in the optic cup of wt (left) and fgf8/3/24-/- (right) embryos at 20ss (dorsal views, anterior to the top). Asterisk in (I): remnant efna5a expression in the optic stalk region. (K) Expression of fgf8 and fgf3 in the dorsal forebrain (fb), and fgf24 in the olfactory placode (olf) in wt embryos at 5ss. Orientation as in (E). (L) Schematic cross-section, illustrating dorsal expression of fgfs and expression of future nasal-temporal retina markers along the dorsal-ventral optic vesicle axis at 10ss. Arrowheads in (E?J): gene expression limits in the optic vesicle and cup. Yellow dotted lines indicate transverse section level. Black and white dotted lines indicate neural tube or optic vesicle/cup outlines. d, dorsal; dn, dorsonasal; dt, dorsotemporal; n, nasal; t, temporal; v, ventral; vn, ventronasal; vt, ventrotemporal. EXPRESSION / LABELING:
PHENOTYPE:
|
|
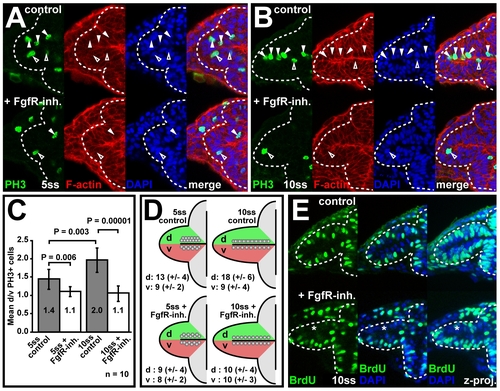
Fgf signaling controls enhanced nasal retina progenitor proliferation. (A and B) Detection of mitotic cells with anti-Phospho Histone H3 (PH3, green), counterstained for cortical F-actin (red) and cell nuclei with DAPI (blue) in the optic vesicle of carrier-treated wt control (top) and FgfR-inh.-treated embryos (bottom) at 5ss (A) and 10ss (B) (filled arrowheads: dorsal leaflet cells, open arrowheads: ventral leaflet cells). (C) Quantification of PH3+ cell numbers in the optic vesicle. Mean ratios of mitotic cells in the dorsal (d) and ventral (v) optic vesicle leaflets (mean d/v PH3+ cells, y-axis) for control (grey) and FgfR-inh.-treated embryos (white) at 5- and 10ss (error bars: standard deviation). (D) Schematic plotting of the mean number of PH3+ cells/optic vesicle dorsal and ventral leaflet (complete anterior-posterior extent of the optic vesicle) into one plane (±standard deviation in brackets). (E) Anti-BrdU staining (green) to detect the proliferation pattern in the optic vesicle at 10 ss in carrier-treated wt control (top) and FgfR-inh.-treated embryos (bottom). Cell nuclei are counterstained with DAPI (blue) (asterisks: reduction in BrdU incorporation, left and middle: single optical sections, right: z-projection). Orientation: cross-sections through one half of the forebrain, dorsal to the top and lateral to the left. Dotted lines in (A, B and E): neural tube boundary. |
|
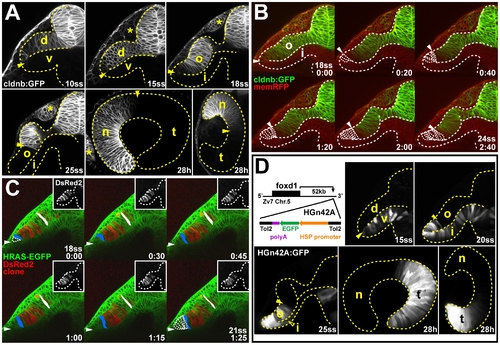
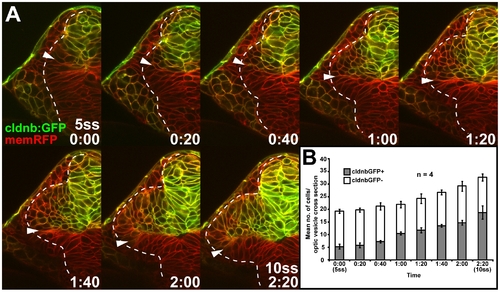
In vivo imaging reveals late movement of prospective temporal retinal cells around the distal ridge of the optic cup. (A) GFP expression (white) in the optic vesicle and optic cup of live Tg(-8.0cldnb:lynGFP)zf106 zebrafish embryos at 10ss, 15ss, 18ss, and 25ss (cross-sections through one half of the forebrain, lateral is to the left) and 28 h (longitudinal section with nasal to the left and horizontal section with nasal to the top). Arrowheads: cldnb:GFP expression limit. (B) Single images from time-lapse analysis of Tg(-8.0cldnb:lynGFP)zf106 expression (green) between 18- (late optic vesicle) and 24ss (early optic cup), colabeled with membrane RFP (memRFP, red), at 20-min intervals. cldnb:GFP- cells moving from the inner optic cup layer, around the distal optic cup ridge are outlined (arrowheads: distal cldnb:GFP expression limit, bottom right: time in hours:minutes). (C) Single images from a time-lapse analysis to track a DsRed2-expressing cell clone (red, insets: single channel) in the outer optic cup layer of a Tg(Bactin:HRAS-EGFP)vu119 host embryo, expressing membrane-targeted GFP (green) between 18- and 21ss, at 30-min intervals. Cells moving from the inner optic cup layer around the distal optic cup ridge are outlined in the first and last image of the series. One cell at the distal limit of the clone (blue) and one cell at the dorsal limit of the clone (white) are pseudocolored (maximal outlines of colored cells and representative apical-basal axis measurements are based on confocal z-stack projections of the complete clone). Arrowheads: distal optic cup ridge, bottom right: time in hours:minutes). (D) Transposon insertion site in HGn42A, 52-kbp downstream of foxd1 on chromosome 5 (Zv7 assembly of the zebrafish genome). GFP expression (white) in the optic vesicle and optic cup of live HGn42A zebrafish embryos at 15ss, 20ss, and 25ss (cross-sections through one half of the forebrain, lateral is to the left) and 28 h (longitudinal section with nasal to the left and horizontal section with nasal to the top) (filled arrowheads: distal GFP expression limit, open arrowhead: GFP expression in inner optic cup layer). d, dorsal optic vesicle leaflet; i, inner optic cup layer; n, nasal; o, outer optic cup layer; t, temporal; v, ventral optic vesicle leaflet. *: olfactory placode. Orientation in (B and C): cross-sections through one half of the forebrain, dorsal to the top and lateral to the left. Dotted lines: neural tube or retina boundaries. EXPRESSION / LABELING:
|
|
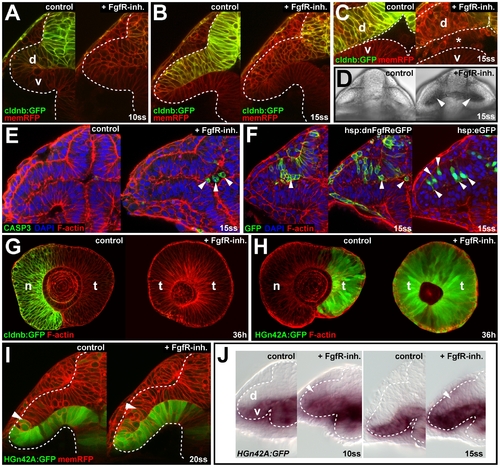
Fgf signaling is required for epithelial integrity of nasal retina progenitors. (A?D) Live images of wt control (left) and FgfR-inh.-treated embryos (right). Tg(-8.0cldnb:lynGFP)zf106 (green) and memRFP colabel (red) at 10ss, 2 h after treatment (hpt) (A), at 15ss, 4 h hpt at low magnification (B), high magnification (C), and in bright field (D) (asterisk in [C] and arrowheads in [D]: delaminated cells in the optic vesicle ventricle). (E) Apoptosis detection with anti-CASP3 (green), counterstained for cortical F-actin (red) and DAPI (blue) in wt control (left) and FgfR-inh.-treated embryo (right) at 15ss, 4 hpt (arrowheads: delaminated cells in the optic vesicle ventricle). (F) Single cells in Tg(hsp70l:dnfgfr1-EGFP)pd1 clones (left two panels) aggregate (left) and protrude apically and are found in the ventricle (middle) at 15ss, 5 h after heat shock, compared to identically treated hsp70l:eGFP control clones (right). Counterstaining: F-actin (red), DAPI (blue). (G) cldnb:GFP expression (green), counterstained for F-actin (red) in the nasal retina of control (left) is absent after FgfR-inh. treatment (right) at 36 h (single, longitudinal confocal sections with nasal to the left). (H) HGn42A:GFP expression (green), counterstained for F-actin (red) in the temporal retina of control (left) is expanded throughout the retina after FgfR-inh. treatment (right) at 36 h (single, longitudinal confocal sections with nasal to the left). (I) Live images of HGn42A:GFP expression (green) in wt control (left) and FgfR-inh.-treated embryos (right) at 20ss, colabeled with membrane-targeted RFP (red) (arrowheads: distal GFP expression limit). (J) HGn42A:GFP transcription detected by GFP in situ hybridization at 10- (left) and 15ss (right) in the ventral optic vesicle leaflet of control embryos is expanded into the dorsal optic vesicle leaflet upon FgfR-inh. treatment (white arrowheads). Orientation in (A?F, I, and J): cross-sections through one half of the forebrain, dorsal to the top and lateral to the left, in (G and H): lateral with nasal/anterior to the left and dorsal to the top. Dotted lines: neural tube boundary. d, dorsal optic vesicle leaflet; n, nasal; t, temporal; v, ventral optic vesicle leaflet. PHENOTYPE:
|
|
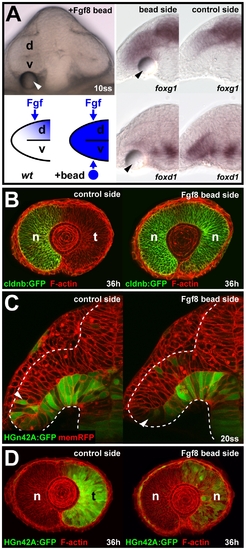
Cell movements into the neural retina can occur independent of Fgf-dependent nasal-temporal patterning. (A) Live embryo at 10ss, 3 h after Fgf8 bead implantation (arrowhead) below the ventral optic vesicle leaflet (top left). Predicted Fgf distribution along the dorsal-ventral axis of the optic vesicle in wt control and after Fgf8 bead implantation (bottom left). Ectopic foxg1 expression (top) and repression of foxd1 (bottom) in the ventral optic vesicle at 10ss, after Fgf8 bead implantation (left) compared to the control side of the same embryo (right). (B) Nasal cldnb:GFP expression (green) at 36 h on the control side (left) and the double-nasal Fgf8 bead implantation side (right) of the same embryo (red: F-actin counterstain). (C) Live images of HGn42A:GFP expression (green), colabeled with membrane-targeted RFP (red) at 20ss after Fgf8 bead implantation (left: control side, right: bead implantation side, arrowheads: distal GFP expression limit). Dotted lines: neural tube boundary. (D) Temporal HGn42A:GFP expression (green) at 36 h on the control side (left) and the double-nasal Fgf8 bead implantation side (right) of the same embryo (red: F-actin counterstain). Orientation in (A and C): cross-sections, dorsal to the top and lateral to the left; in (B and D): lateral with nasal/anterior to the left and dorsal to the top. d, dorsal optic vesicle leaflet; n, nasal; t, temporal; v, ventral optic vesicle leaflet. |
|
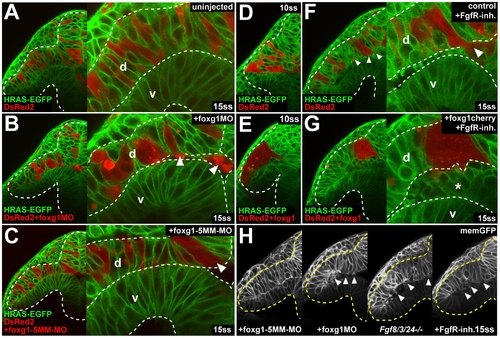
Foxg1 is required for cell cohesion in the optic vesicle. (A-C) Live images of a DsRed2-expressing cell clone from a noninjected control donor (A), a foxg1-morpholino-injected donor (B), and a foxg1-mismatch-control-morpholino-injected donor in a Tg(Bactin:HRAS-EGFP)vu119 host embryo (C) at 15ss (right: high magnification view of the dorsal and ventral leaflet in the region of the proximal optic vesicle, arrowheads: delaminating cells). (D and E) Live images of a DsRed2-expressing cell clone from a noninjected control donor (D) and a foxg1-RNA-injected donor (E) in a Tg(Bactin:HRAS-EGFP)vu119 host embryo at 10ss. (F and G) Live images of a DsRed2-expressing cell clone from a noninjected control donor (F) and a foxg1-injected donor (G) in a Tg(Bactin:HRAS-EGFP)vu119 host embryo in the presence of FgfR-inhibitor at 15ss (right: high magnification view of the dorsal and ventral leaflet in the region of the proximal optic vesicle, arrowheads in [F] and asterisk in [G]: delaminating cells). (H) Live images of cell delamination (arrowheads) in a foxg1-mismatch-control-morpholino-injected embryo, a foxg1-morpholino-injected embryo, an fgf8/3/24-/- embryo, and an embryo after FgfR.-inh. treatment at the 15ss stage, coinjected with memGFP RNA (from left to right). Orientation: cross-sections, dorsal to the top and lateral to the left. Dotted lines: neural tube boundary. d, dorsal optic vesicle leaflet; v, ventral optic vesicle leaflet. PHENOTYPE:
|
|
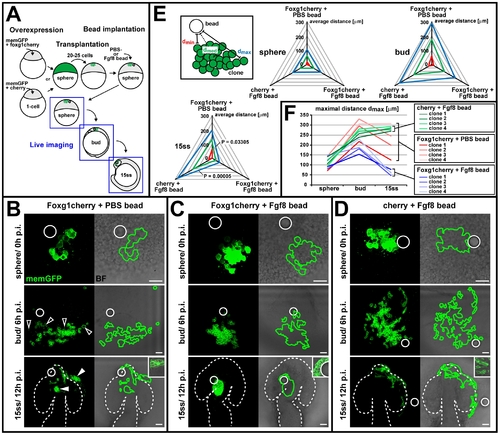
Foxg1 mediates cell cohesion in an Fgf-dependent manner. (A) Schematic of the overexpression-cell-transplantation-bead-implantation assay to test for an Fgf-dependent effect of foxg1 on cell cohesion by monitoring lateral cell spreading in live embryos. (B?D) Live images of horizontal cell spreading at sphere (0 h postimplantation [p.i.], top), bud (6 h postimplantation, middle), and 15ss (12 h postimplantation, bottom) from a representative clone overexpressing foxg1cherry in the presence of a PBS control bead (B), a clone overexpressing foxg1cherry in the presence of an Fgf8 bead (C), and a clone overexpressing cherry in the presence of an Fgf8 bead (D). Left: memGFP image (confocal z-projection), right: clone outline (green line), derived from memGFP image, superimposed with a bright-field (BF) image, showing bead localization (white circle). Dotted outlines: neural tube border. Insets in lower panels show cell morphology (bright-field image superimposed with a single confocal section of memGFP). Open arrowheads in (B): abnormal cells not included in tracking the clone outline. White arrowheads in (B): autonomous clustering of foxg1-overexpressing cells in the olfactory placode, head mesenchyme, and telencephalon. Scale bar: 50 μm. (E) Quantification of lateral cell spreading relative to the site of bead implantation. The maximal cell distance (dmax, blue) and the minimal distance from the bead surface (dmin, red) was measured (on projected z-stacks) and the median distance (dmed, green) calculated on the basis of dmax and dmin for four representative clones at sphere, bud, and 15ss. Radial plots of average dmax, dmin, and dmed values (in micrometers) at sphere, bud, and 15ss, (n = 4/stage) for foxg1cherry overexpression in the presence of a PBS control bead (top axis), foxg1cherry overexpression in the presence of an Fgf8 bead (right axis), and cherry overexpression in the presence of an Fgf8 bead (left axis). p-Values for 15ss stage derived by a paired, two-tailed t-test. (F) Line plots of single dmax values (in micrometers) over time (sphere, bud, 15ss) for four representative clones overexpressing foxg1cherry in the presence of a PBS control bead (red), overexpressing foxg1cherry in the presence of a Fgf8 bead (blue), and overexpressing cherry in the presence of a Fgf8 bead (green). Brackets indicate dmax value range at 15ss. |
|
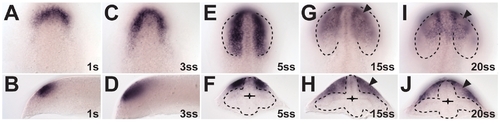
fgf24 expression in the olfactory placode at optic vesicle and cup stages. (A and B) Onset of expression, anterior to the forebrain (1s). (C?F) Posterior expansion along the hinge between dorsal optic vesicle leaflet and dorsal forebrain (3- and 5ss). (G?J) Lateral spreading, anterior condensation and olfactory pit preformation (see fgf24-nonexpressing cells, arrowheads) at 15- and 25ss. (A, C, E, G, and I) Dorsal view, anterior to the top. (B and D) Lateral view, anterior to the left. (F, H, and J) Cross-section, dorsal to the top. Dotted outlines: optic vesicle and cup boundary (E, G, and I) or neural tube boundary (F, H, and J). |
|
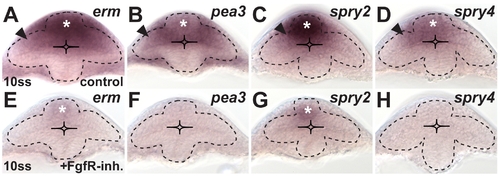
Target gene expression shows active Fgf signaling in the dorsal optic vesicle leaflet. Expression of the Fgf target genes erm, pea3, spry2, and spry4 at the 10ss stage in the optic vesicle of control embryos (A?D) and embryos after FgfR-inhibitor treatment (E?H). (A) erm is strongly expressed in the dorsal forebrain (asterisk) and the dorsal optic vesicle leaflet (arrowhead) of the control. (E) Remnant erm expression is only found in the dorsal forebrain in inhibitor treated embryos (asterisk). (B) pea3 is weakly expressed in the dorsal forebrain (asterisk) but stronger in the proximal part of the dorsal optic vesicle leaflet (arrowhead) of the control. (F) Inhibitor-treated embryos show no pea3 expression. (C) spry2 is strongly expressed in the dorsal forebrain (asterisk) and the proximal part of the dorsal optic vesicle leaflet (arrowhead) of the control. (G) Remnant spry2 expression is only found in the dorsal forebrain of inhibitor treated embryos (asterisk). (D) spry4 is weakly expressed in the dorsal forebrain (asterisk) and in the proximal part of the dorsal optic vesicle leaflet (arrowhead) of the control. (H) Inhibitor-treated embryos show no spry4 expression. All images are cross-sections, dorsal to the top; dotted lines: neural tube boundary. |
|
In vivo imaging of clnb:GFP expression between the 5- and 10ss stages. (A) Single images from a confocal time-lapse series of cldnb:GFP expression (green) colabeled with membrane-targeted RFP (memRFP, red), captured at 20-min intervals (cross-section through one half of the forebrain, lateral to the left and dorsal to the top, bottom right: time in hours:minutes). (B) Mean number of cldnb:GFP-expressing (cldnbGFP+, grey) and nonexpressing (cldnbGFP-, white) cells in single cross-sections (captured at a 50?70-μm depth from anterior optic vesicle tip) through the optic vesicle between 5- and 10ss (error bar: standard deviation). |
|
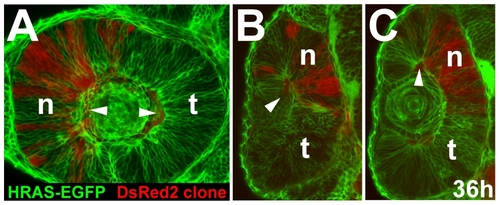
Nasal restriction of cells from the outer optic cup layer. The DsRed2 cell clone form Figure 3C is restricted to the nasal retina of the HRAS-EGFP host at 36 h. (A) lateral view, (B) optical cross-section at a ventral (B) and medial (C) level along the nasal-temporal axis (arrowheads: autofluorescent blood vessels). n, nasal; t, temporal. |
|
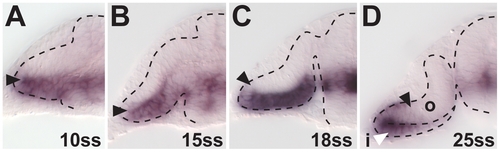
foxd1 expression in temporal retina progenitors during optic cup formation. (A and B) At 10ss (A) and 15ss (B), foxd1 expression is confined to the ventral optic vesicle leaflet. (C) At 18ss, the first foxd1-positive cells are found at the distal part of the forming outer optic cup layer, indicating the onset of temporal retina progenitor movement into the future neural retina. The whole ventral leaflet is expressing foxd1. (D) At 25ss, foxd1 expression is found in the ventral part of the outer optic cup layer, indicating continued movement. Only the distal part of the inner optic cup layer contains foxd1-expressing cells (white arrowhead), indicating continued movement of temporal progenitors out of this region (black arrowheads: distal/dorsal gene expression limit, dotted lines: neural tube boundary. All images are cross-sections. |
|
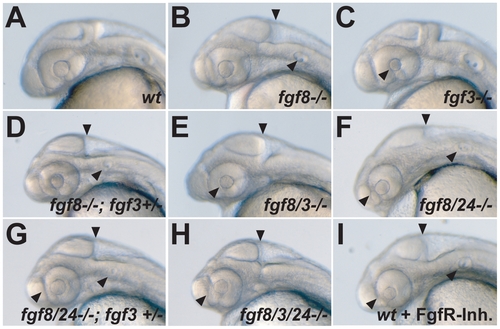
Live phenotype of Fgf8/3/24 mutant embryos at 28 h. (A) wt control embryo, (B) fgf8-/- mutant with reduced ear and lacking cerebellum (arrowheads), (C) fgf3-/- mutant with small lens (arrowhead), (D) fgf8-/-; fgf3+/- transheterozygous embryo, lacking the cerebellum and with strongly reduced ear (arrowheads), (E) fgf8/3-/- double-mutant, lacking cerebellum and ear and with small lens (arrowheads), (F) fgf8/24-/- double mutant, showing nasally tilted eye position, lack of the cerebellum, and reduction of the ear, (G) fgf8-/-; fgf3+/- transheterozygous embryo, injected with fgf24 MO (fgf8/24-/-; fgf3+/-), showing nasally tilted eye position, lack of the cerebellum, and strong reduction of the ear (arrowheads), (H) fgf8/3-/- double mutant injected with fgf24 MO (fgf8/3/24-/-), showing nasally tilted eye position, lack of the cerebellum and ear (arrowheads), and (I) wt embryos treated with FgfR-inh. (wt+FgfR-Inh.), showing nasally tilted eye position, lack of cerebellum, and reduced ear (arrowheads). |
|
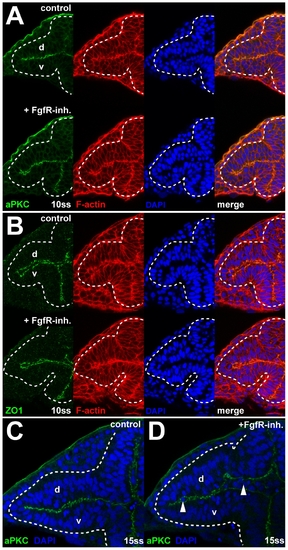
Apical-basal cell polarity after FgfR inhibition. (A and B) Normal localization of the apical membrane markers aPKC (A) and ZO1 (B) at 10ss, after FgfR-inh. treatment (bottom panels) compared to control embryos (top panels). (C and D) Cell delamination in the optic vesicle after FgfR-inh. treatment (D) compared to control (C) at 15ss. Sites of delamination (arrowheads in [D]) correspond to regions where apical membrane contact between dorsal and ventral leaflet (revealed by staining for aPKC, green) is lost. Images are transverse sections, counterstained with DAPI (blue) and for F-actin (red), dorsal to the top, dotted lines: neural tube boundary. d, dorsal optic vesicle leaflet; v, ventral optic vesicle leaflet. |
|
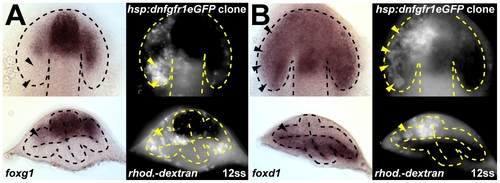
Effect of clonal dnFgfR1 overexpression on foxg1 and foxd1 in the evaginating optic vesicle. Clonal dnFgfr1 overexpression by transplantation and heat-shock induction of rhodamine-dextran lineage-labeled Tg (hsp70l:dnfgfr1-EGFP)pd1 cells. dnFgfr1 overexpression in the dorsal optic vesicle leaflet (arrowheads) represses foxg1 expression (A) and leads to ectopic foxd1 expression (B) at 12ss. Heat shocks were given at the onset of optic vesicle evagination (1-3ss). Top panels: dorsal views, bottom panels: cross-sections with dorsal to the top, left panels: bright field, right panels: fluorescent lineage label, and dotted lines: neural tube boundary (bottom panels) or optic vesicle boundary (top panels). |
|
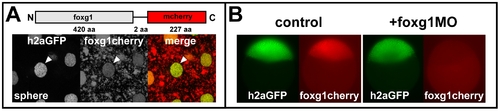
Foxg1 morpholino knockdown. (A) Live image of Foxg1-cherry fusion protein expression (red) in the animal pole blastoderm in a Tg (h2afv:GFP)kca66 embryo (green) at sphere stage shows nuclear (arrowhead) and cytoplasmic localization of the protein. (B) Compared to a noninjected control (left), injection of foxg1 morpholino (foxg1MO, right) results in complete and specific depletion of the fluorescent foxg1cherry signal (red) compared to Tg (h2afv:GFP)kca66 (h2aGFP, green) in live embryos at sphere stage (lateral views, animal to the top). |