- Title
-
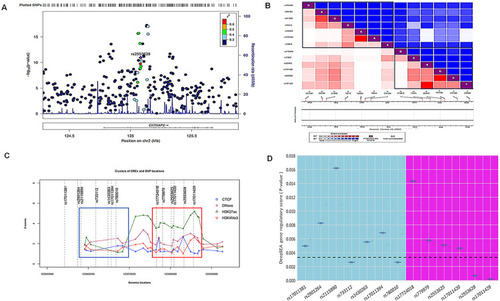
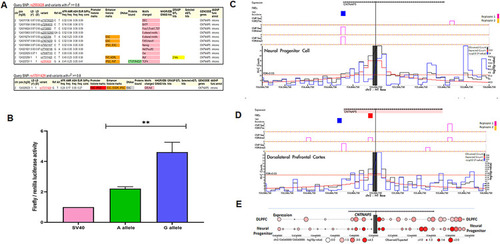
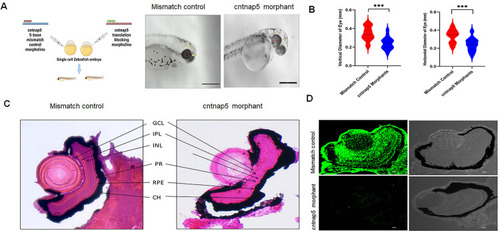
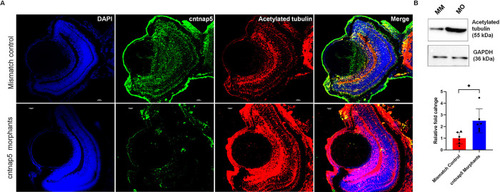
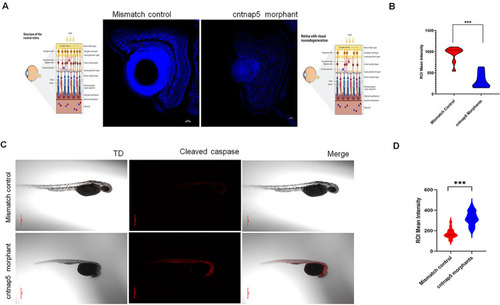
Functional investigation suggests CNTNAP5 involvement in glaucomatous neurodegeneration obtained from a GWAS in primary angle closure glaucoma
- Authors
- Chakraborty, S., Sarma, J., Roy, S.S., Mitra, S., Bagchi, S., Das, S., Saha, S., Mahapatra, S., Bhattacharjee, S., Maulik, M., Acharya, M.
- Source
- Full text @ PLoS Genet.
|
|
|
|
|
|
|
|
|
PHENOTYPE:
|
|
PHENOTYPE:
|