- Title
-
ALS-linked VapB P56S mutation alters neuronal mitochondrial turnover at the synapse
- Authors
- Wong, H.C., Lang, A.E., Stein, C., Drerup, C.M.
- Source
- Full text @ J. Neurosci.
|
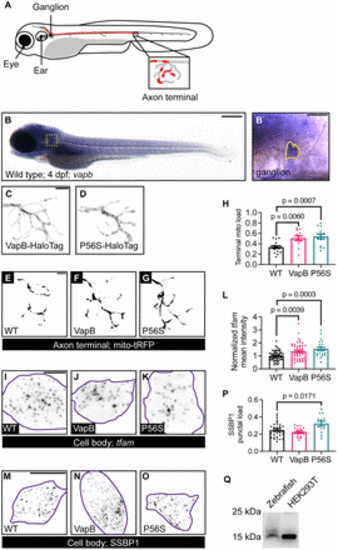
VapB regulates mitochondrial density in the axon terminal. A, Schematic of the pLL in the zebrafish larva. Neuronal cell bodies reside in the pLL ganglion and extend axons down the trunk. Single axon in red. Axon terminal in inset. A fraction of the axon terminal is occupied by mitochondria (red in inset). B, In situ hybridization of vapb at 4 dpf. vapb is enriched in pLL neuron cell bodies. pLL ganglion boxed in B, outlined in B`. C, D, Representative images of pLL axon terminals expressing VapB (C) or VapBP56S (P56S; D) tagged with a HaloTag. E?G, Representative images of mitochondria visualized with matrix-localized TagRFP (tRFP) in axon terminal of WT (E), VapB transgenic (F), or VapBP56S transgenic (G) larvae at 4 dpf. (H) Quantification of mitochondrial load (mitochondrial area/axon terminal area) with manipulation of VapB. I?K, Images of tfam HCR RNA FISH from single pLL neuron (purple outline) coexpressing GFP (I), VapB (J), or VapBP56S (K). L, Quantification of tfam mean fluorescence intensity normalized to WT. M?O, Representative images of SSBP1 immunostaining in single pLL neurons (purple outline) coexpressing GFP (M), VapB (N), or VapBP56S (O). P, Quantification of the SSBP1 puncta number normalized to the cell body area. Q, Anti-SSBP1 Western blot of WT larval protein extract and HEK293T cell lysate. Scale bar: 200 ?m in B; 50 ?m in B`; 5 ?m in C and E; 10 ?m in I and M. Each data point represents the average calculated from an individual animal. All data represented as mean ± SEM. ANOVA with Dunnett's post hoc contrasts in H, L, and P. |
|
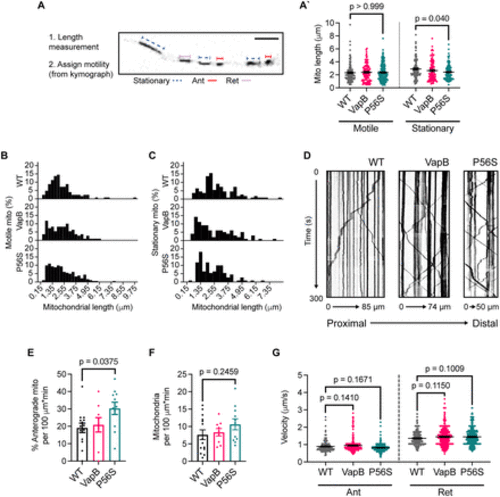
VapBP56S expression decreases mitochondrial length and increases mitochondrial anterograde transport. A, Average length of motile (left) and stationary (right) mitochondria with VapB and VapBP56S transgenic expression. Schematic of mitochondrial length measurement in A. The long axis of mitochondria was measured as the length. B, C, Frequency distribution histograms of mitochondrial lengths for motile (B) and stationary mitochondria (C) in pLL axons. The number of mitochondria and animals analyzed: WT, 124, 14; VapB, 110, 9; and P56S, 182, 11 for motile mitochondria (B), and WT, 109, 13; VapB, 98, 9; and P56S, 110, 11 for stationary mitochondria (C). D, Example kymographs of mitochondrial transport in WT, VapB transgenic, and VapBP56S transgenic backgrounds. E, F, Quantification of the number of mitochondria moving in the anterograde direction (E) and total number of mitochondria (F) in pLL axons. Each data point represents the average calculated from an individual animal. G, Analysis of average anterograde (Ant; left) and retrograde (Ret; right) transport velocity. The number of mitochondria and animals analyzed: WT, 161, 17; VapB, 331, 9; and P56S, 625, 13 for anterograde transport, and WT, 262, 16; VapB, 427, 8; and P56S, 471, 12 for retrograde transport. All data represented as mean ± SEM. Kruskal?Wallis test with Dunn's post hoc contrasts in A and G. ANOVA with Dunnett's post hoc contrasts in E and F. |
|
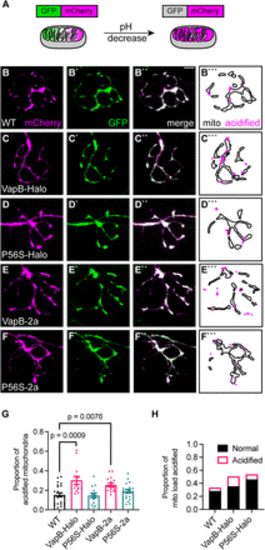
VapB expression increases mitophagy in the axon terminal. A, Schematic of mitophagy indicator mito-GFP-mCherry localization and fluorescence in the mitochondrion. In low pH, such as in a lysosome, the GFP is quenched. B?D, Representative images of axon terminals expressing the mitophagy indicator in WT (B), VapB-HaloTag transgenic (VapB-Halo; C), VapBP56S-HaloTag transgenic (P56S-Halo; D), VapB-p2a-TagBFP transgenic (VapB-2a; E), and VapBP56S-p2a-TagBFP transgenic (P56S-2a; F). Distribution of acidified (magenta) and unacidified (outline) mitochondria is illustrated (B```?F```). G, Proportion of acidified mitochondria relative to the total mitochondrial population. H, Proportion of mitochondrial load acidified (mCherry-only mitochondrial area/average mitochondrial axon terminal load derived from Fig. 1H). Scale bar, 5 ?m in B``. Each data point for G represents the average calculated from an individual animal. All data represented as mean ± SEM. ANOVA with Dunnett's post hoc contrasts in G. |
|
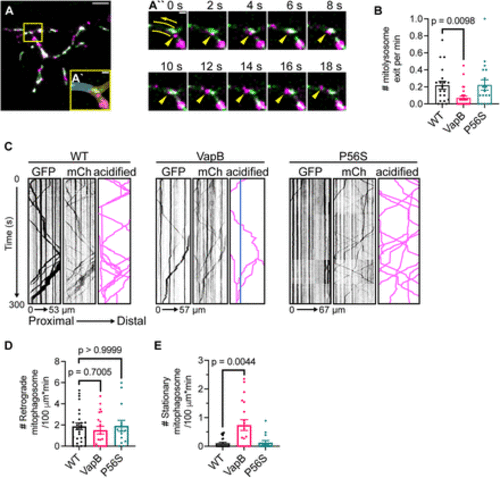
Exogenous VapB expression stalls mitophagosome transport. A, Representative image of mitophagosome exits from a WT axon terminal. Branchpoint connecting axon to axon terminal boxed and magnified in A` with axon highlighted in blue and axon terminal in yellow. The 18 s excerpt of Movie 1 axon branchpoint time lapse shows mitophagosome (yellow arrowhead) exit from the example axon terminal (A``). The arrow denotes direction of retrograde transport toward cell body, two lines border the path of mitophagosome travel. See Movie 1. B, Quantification of mitophagosome exit event frequency calculated from 20 min time-lapse imaging sessions. C, Example kymographs from axons of WT, VapB transgenic, and VapBP56S transgenic expressing the mitophagy indicator showing transport of unacidified (GFP) and all mitochondria (mCh). Schematic traces of acidified mitochondrial transport are illustrated on the right. Stationary acidified mitochondria traced in blue. D, E, Number of retrograde (D) and stationary (E) acidified mitochondria (mCherry only) in pLL axons expressing the mitophagy indicator. Scale bar: 5 ?m in A, 1 ?m in A` and A``. Each data point for B, D, and E represents the average calculated from an individual animal. All data represented as mean ± SEM. Kruskal?Wallis with Dunn's post hoc contrasts in B, D, and E. |
|
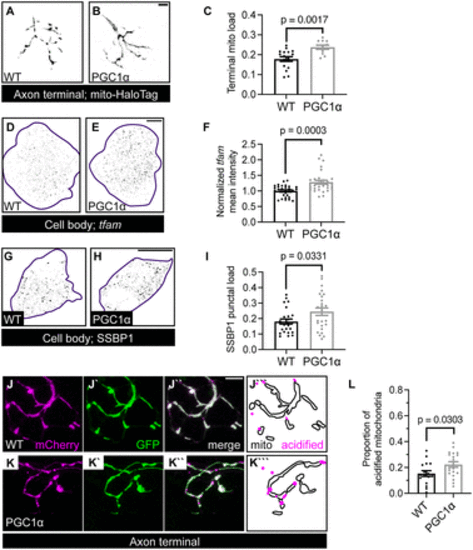
PGC1? regulates mitochondrial density and mitophagy in the axon terminal. A, B, Representative images of mitochondria, visualized with HaloTag localized to the mitochondrial outer membrane via a Tomm20 signal sequence, in axon terminals of a WT (A) and PGC1? transgenic (B) at 4 dpf. C, Quantification of axon terminal mitochondrial load in PGC1? transgenic. D, E, Representative images of HCR RNA FISH labeling tfam mRNA in the pLL ganglion (purple outline) of WT (D) and PGC1? transgenic (E). F, Quantification of tfam mean fluorescence intensity normalized to WT. G, H Images of SSBP1 immunostaining in single pLL neurons (purple outline) in WT (G) and PGC1? transgenic (H). I, Quantification of SSBP1 puncta number normalized to the cell body area. J, K, pLL axon terminal of WT (J) and PGC1? transgenic (K) expressing the mitophagy indicator. Distribution of acidified (magenta) and unacidified (outline) mitochondria is illustrated (J```, K```). L, Quantification of axon terminal mitophagy in PGC1? transgenic. Scale bar, 5 ?m in B and J``; 10 ?m in E and H. Each data point for C, F, I, and L represents the average calculated from an individual animal. All data represented as mean ± SEM. Student's t test in C, I, and L. Mann?Whitney U test in F. |
|
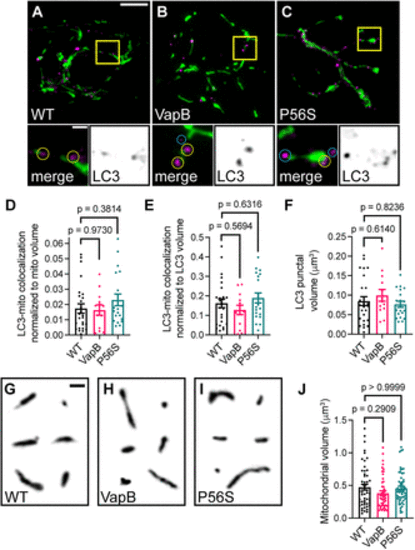
Mitochondrial association with autophagosome membranes is not disrupted by VapB or VapBP56S expression. ??C, Deconvolved images of pLL axon terminals coexpressing mitochondrially localized GFP (green) and RFP-LC3 to label autophagosomes (RFP in magenta in merge, black on the right inset) in WT (A), VapB transgenic (B), or VapBP56S transgenic (C). Inset below; yellow circles denote mitochondria-associated autophagosome; blue circles denote unassociated autophagosomes. D, Proportion of the mitochondrial volume colocalized with autophagosomes normalized to mitochondrial volume. E, Proportion of the mitochondrial volume colocalized with autophagosomes normalized to autophagosome volume. F, Average autophagosome volume in the axon terminal. G?I, Example mitochondria from deconvolved images of axon terminals of WT (G), VapB transgenic (H), or VapBP56S transgenic (I). J, Average mitochondrial volume in the axon terminal. Scale bar: 5 ?m in A; 1 ?m in A inset and G. Each data point for D?F represents the average calculated from an individual animal. Data points in J represent 158 mitochondria from WT, 10; VapB, 8; P56S, 8 animals per group. All data represented as mean ± SEM. ANOVA with Dunnett's post hoc contrasts in D, E, and F. Kruskal?Wallis with Dunn's post hoc contrasts in J. |
|
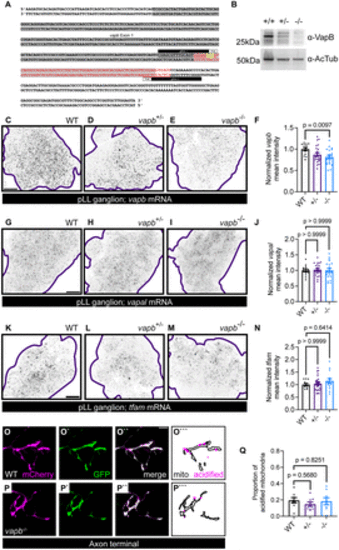
vapb knock-out does not phenocopy VapBP56S or WT VapB transgenic expression. A, vapb exon 1 with deletion in the vapb mutant in red. Guide RNAs used to generate line are shown. This mutation eliminates the start site for protein translation (shown in yellow). B, Anti-VapB Western blot of whole larval protein extracts from WT, heterozygous siblings, and vapb mutants. C?E, Images of the pLL ganglion (outlined) of HCR RNA FISH labeling vapb mRNA in WT (C), heterozygous (D; vapb+/?), or vapb mutant (E; vapb?/?) animals. F, Quantification of vapb mRNA mean fluorescence intensity normalized to WT. G?I, Images of the pLL ganglion (outlined) of HCR RNA FISH labeling the vapb homolog vapal. WT (G), heterozygous (H; vapb+/?), and vapb mutant (I; vapb?/?) animals are shown. J, Quantification of vapal mRNA mean fluorescence intensity normalized to WT. K?M, Images of HCR RNA FISH labeling tfam mRNA in the pLL ganglion (outlined) in WT (K), heterozygous (L; vapb+/?), or vapb mutant (M; vapb?/?) animals. N, Quantification of tfam mean fluorescence intensity normalized to WT. O, P, Representative images of axon terminals expressing the mitophagy indicator in WT (O) and vapb?/? animals (P). Distribution of acidified (magenta) and unacidified (outline) mitochondria illustrated (O```?P```). Q, Proportion of acidified mitochondrial area relative to total mitochondrial population area. Kruskal?Wallis with Dunn's post hoc contrasts for F, J. ANOVA with Dunnett's post hoc contrasts in N, Q. Scale bar, 10 ?m in C, G, and K, 5 ?m in O``. Each data point for F, J, N, and Q represents the average calculated from an individual animal. All data represented as mean ± SEM. |
|
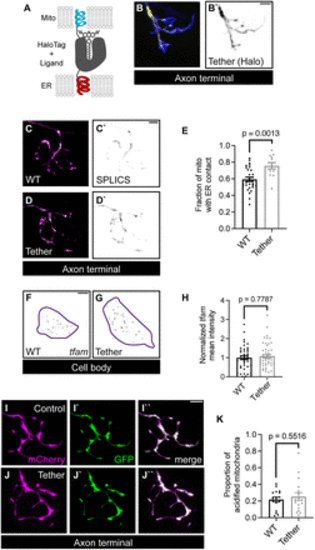
Mitochondrial biogenesis and mitophagy regulation are independent of ER?mitochondrial tethering. A, Schematic of synthetic ER?mitochondrial tether. This tether is composed of the mitochondrial localization sequence from Tomm20 and the ER localization sequence from Sec61? linked with a HaloTag sequence. B, Expression of the synthetic tether (yellow in B, black in B`) in a pLL axon terminal. GFP labels the cytoplasm (blue). C, D, SPLICS signal (green on top, black on bottom) visualizing mitochondria (magenta) proximity to the ER membrane. E, Quantification of SPLICS-positive mitochondrial area in pLL axon terminals. F, G, A representative image of HCR RNA FISH labeled tfam fluorescence in single pLL neuron (purple outline) in individual pLL neurons expressing cell-fill GFP (F) or synthetic tether (G). H, Quantification of mean tfam fluorescence intensity normalized to WT levels. I, J, A representative image of axon terminal GFP and mCherry signals in pLL neuron expressing the mitophagy indicator in WT (I) or the synthetic tether transgenic (J). K, Quantification of the proportion of acidified mitochondria with tether expression. Student's t test for E and K. Mann?Whitney U test for H. Scale bar, 5 ?m. Each data point for E, H, and K represents the average calculated from an individual animal. All data represented as mean ± SEM. |