- Title
-
METTL3-dependent m6A modification of PSEN1 mRNA regulates craniofacial development through the Wnt/β-catenin signaling pathway
- Authors
- Ma, L., Zhou, X., Yao, S., Zhang, X., Mao, J., Vona, B., Fan, L., Lou, S., Li, D., Wang, L., Pan, Y.
- Source
- Full text @ Cell Death Dis.
|
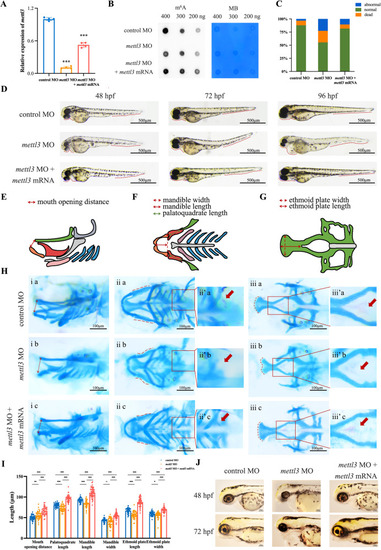
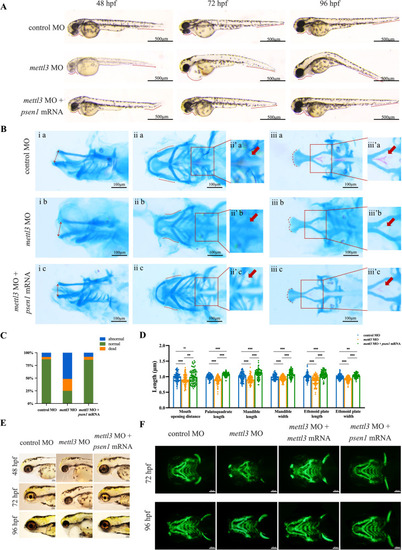
Phenotypes of the zebrafish. |
|
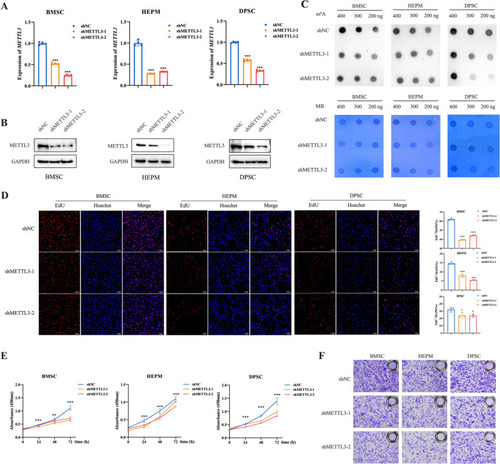
METTL3 knockdown significantly suppresses cell proliferation and migration in vitro. |
|
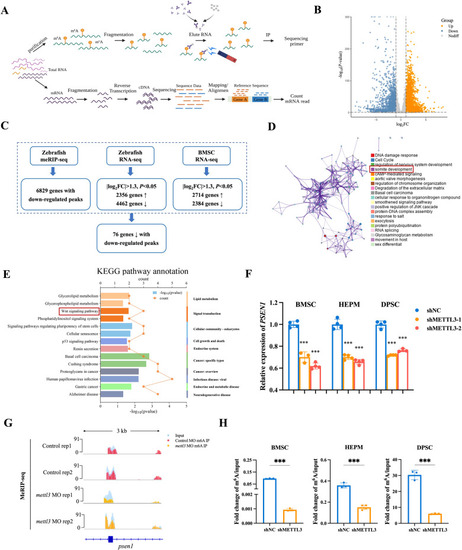
Identification of METTL3 targets via MeRIP-seq and RNA-seq. |
|
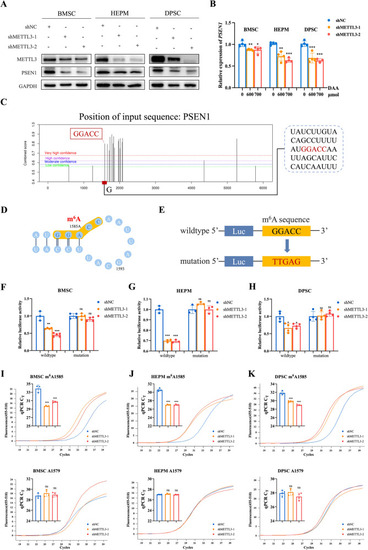
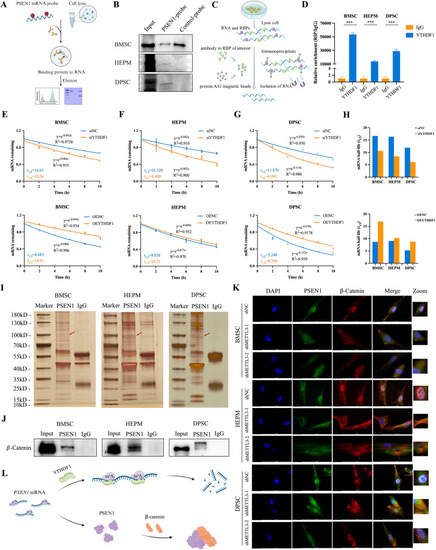
PSEN1 is modulated by METTL3-mediated m6A RNA methylation. |
|
|
|
PSEN1 is specifically recognized by YTHDF1 and directly interacts with β-catenin. |
|
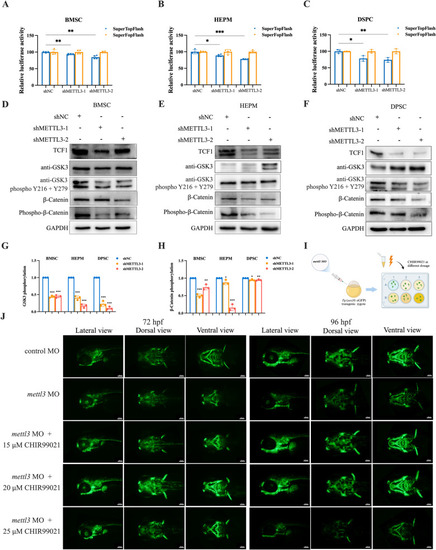
METTL3 deficiency inhibits Wnt/β-catenin signaling and Wnt/β-catenin activation partially alleviates the phenotypes of |