- Title
-
Latent developmental potential to form limb-like skeletal structures in zebrafish
- Authors
- Hawkins, M.B., Henke, K., Harris, M.P.
- Source
- Full text @ Cell
|
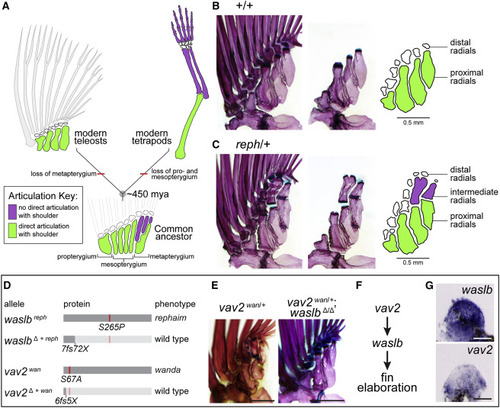
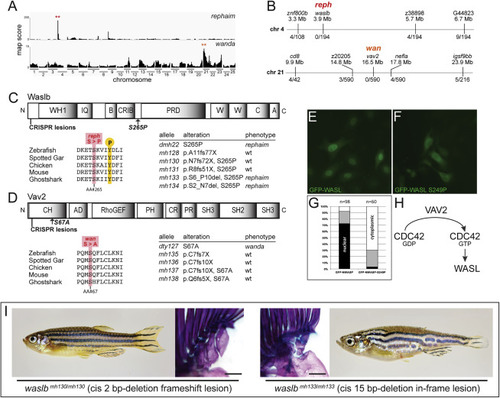
Figure 1. Gain-of-function mutations reveal capacity for limb-like development in fins (A) From a common ancestor, teleosts and tetrapods evolved divergent appendage patterns (see also Figure S1). (B and C) Adult pectoral fin skeletons shown intact (left); fin rays and distal radials removed (center); schematized (right) (see also Figure S2). (B) Wild-type pectoral skeleton. (C) Heterozygous reph mutant pectoral skeleton. (D) waslb and vav2 alleles causing mutant phenotypes; ?+ reph (mh130), and ?+ wan (mh138) rescue wild-type patterning (see also Figure S3). (E) Epistasis analysis showing suppression of wan/vav2 phenotypes after loss of waslb function (see also Figure S4). (F) Genetic pathway of vav2, waslb, and fin elaboration. (G) Expression of waslb and vav2 in 48 hpf pectoral fin buds. Anterior to left, distal to top; scale bars (B, C, E) 500 ?m, (G) 50 ?m. |
|
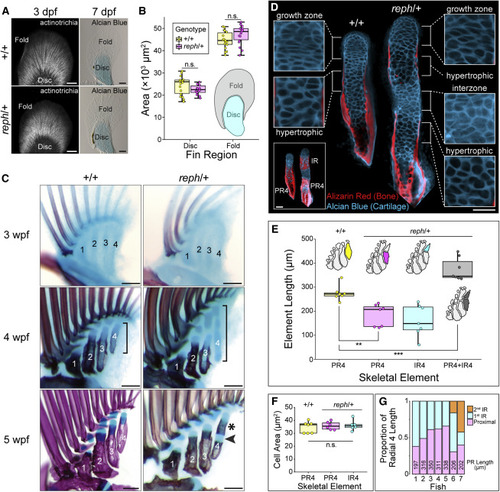
Figure 2. Novel intermediate radial elements arise by segmentation of a common precursor (A) The fin fold forms normally in reph/+ mutants. Immunolabeling of actinotrichia fibers at 3 dpf reveals no gross defects in the fin fold (n = 10). Alcian blue staining of cartilage at 7 dpf shows similar relative proportion of fin fold and endoskeletal disc size in wild-type and reph/+ animals. (B) Endoskeletal disc area at 7 dpf is not significantly different between wild-type and reph/+ fins (n = 18 each genotype, Welch?s Two Sample t test, t = ?1.7605, p > 0.09). Fin fold area is also not significantly different (n = 18 each genotype, Welch?s Two Sample t test, t = 1.8918, p > 0.06). (C) Fin ontogeny in wild-type and reph/+ animals. Initial separation of the endoskeletal disc into the four proximal radial precursors is underway at 3 wpf. Proximal mineralization of the radials is evident at 4 wpf, while posterior radial cartilages are elongated in reph/+ mutants (bracket). At 5 wpf, wild-type proximal radials exhibit continuous zones of mineralization, while reph/+ mutants have a secondary bone collar (asterisk) separated from the proximal ossification by cartilage (arrowhead). (D) Elongated radials form interzones at sites of future segmentation. Confocal sections of wild-type and reph/+ radials at 5 wpf, bottom left inset shows full confocal projection, other insets magnify cartilage regions. (E) Quantification of radial element 4 lengths in wild-type and mutant animals at 5 wpf. Total reph/+ radials are significantly longer than wild-type PR4 (n = 7 each genotype, Welch?s Two Sample t test, t = 4.64, p = 8 × 10?4) while reph/+ PR4 elements are significantly shorter than wild-type elements (n = 7 each genotype, Welch?s Two Sample t test, t = ?4.36, p = 1 × 10?3). (F) Quantification of average hypertrophic chondrocyte size at 5 wpf reveals no significant difference between wild-type and reph/+ elements (n = 7 each element type, one-way ANOVA, F = 0.319, p = 0.732). (G) Segmentation position along radial 4 is variable among reph/+ mutants. Absolute size of PR4 provided for each fish. Anterior to left, distal to top in all micrographs; PR4, proximal radial 4; IR4, intermediate radial 4; n.s., not significant; scale bars (A) 50 ?m, (C) 125 ?m, (D) 50 ?m. PHENOTYPE:
|
|
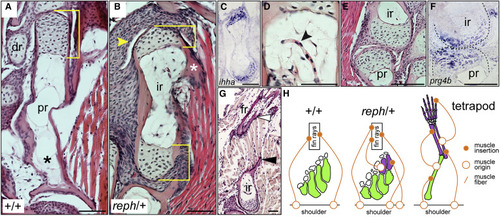
Figure 3. Intermediate radials are long bones morphologically integrated into the fin (A?G) Histology of adult fins. (A) Wild-type proximal radial with a single distal epiphysis (bracket). (B) reph/+ intermediate radial with dual epiphyses (brackets), a muscle insertion point (asterisk), and distal synovial cavity (arrowhead). (C) ihha expression in epiphyses of an intermediate radial. (D) Blood vessel invading an intermediate radial (black arrowhead). (E) Synovial joint between intermediate and proximal radials. (F) prg4b expression in the proximal-intermediate radial joint. (G) Muscle fibers originating from process on intermediate radial (black arrowhead) and inserting on the dermal fin rays (white arrowhead). (H) Diagram of muscle attachment in the tetrapod limb versus wild-type and reph/+ zebrafish pectoral fins. Anterior to left, distal to top; dr, distal radial; fr, fin ray; ir, intermediate radial; pr, proximal radial; scale bars 50 ?m. |
|
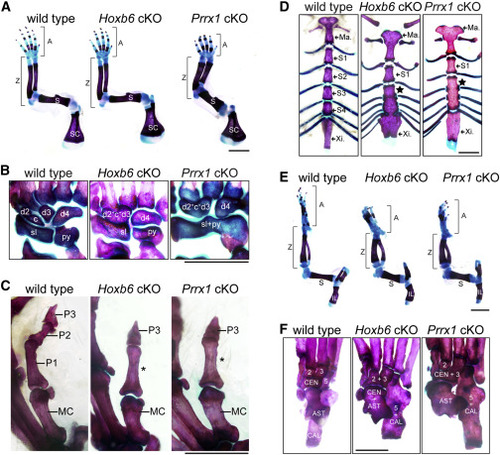
Figure 4. Conditional knockouts reveal Wasl is required for normal skeletal patterning in mice (A) Forelimbs of adult wild-type and conditional Wasl knockout (cKO) mice. Prrx1 cKO mice exhibit shortened and thickened long bones. (B) Conditional Wasl inactivation causes carpal coalition. (C) Phalanges of Digit V. Conditional knockout animals lack a discernable intermediate phalanx (P2) and instead have a single phalanx (asterisk) between the metacarpal (MC) and distal phalanx (P3). (D) Fused sternebrae (star) in conditional Wasl knockout mice. (E) Hindlimbs of adult wild-type and conditional Wasl knockout mice. Hoxb6 and Prrx1 cKO mice exhibit shortened and thickened long bones of the leg. (F) Ankle bone fusions in conditional Wasl knockout mice. See also Table S1. Distal to top in (A), (B), (C), (E), (F); rostral to top in (D); anterior to left in (B), (C), and (F). 2, distal tarsal 2; 3, distal tarsal 3; 5, distal tarsal 5; AST, astragalus; A, autopod; CAL, calcaneus; CEN, centrale; c, central carpal; d#, distal carpal number; IL, ilium; Ma, manubrium; MC, metacarpal; py, pyramidal; P#, phalanx number; S, stylopod; SC, scapula; sl, scapholunate; S#, sternebrae number; Xi, xiphoid process; Z, zeugopod; scale bars 2 mm. |
|
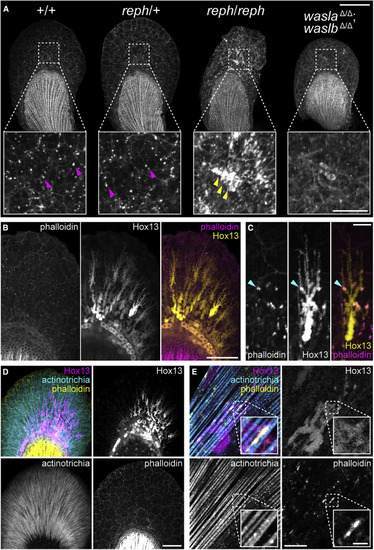
Figure 5. F-actin formation in the early fin fold is affected in wasl mutants and is associated with Hox13-positive cells and the actinotrichia (A) 3 dpf phalloidin-labeled pectoral fins showing modified F-actin in reph homozygous mutants and wasla; waslb double mutants. reph homozygotes develop large F-actin aggregates and dysmorphic fin folds, and wasla; waslb double mutants display a more dispersed F-actin network. (B) F-actin foci colocalize with the extensions of Hox13-positive cells in the fin fold marked with the Tg(2P?epi:eGFP) reporter transgene. (C) A single Tg(2P?epi:eGFP) Hox13-positive cell showing F-actin colocalization. (D) Triple labeling demonstrates colocalization of F-actin foci, Hox13-positive cells expressing the Tg(mInta-11:mCherry) transgene, and actinotrichia in the fin fold. (E) A single Tg(mInta-11:mCherry) Hox13-positive cell showing colocalization with actinotrichia fibrils and F-actin. Anterior to left, distal to top in all panels; scale bars (A) 100 ?m (25 ?m inset), (B, D) 50 ?m, (C, E) 10 ?m (2 ?m inset in E). PHENOTYPE:
|
|
Figure 6(. Expression of hoxa11b is upregulated in reph mutants and is required for the formation of intermediate radials (A?F) Genetic interaction between waslb and hox genes in zebrafish fin patterning. (A) Wild-type pectoral fin. (B) waslbreph/+ mutants forming intermediate radials (arrowhead; n = 9, 89%). (C) hoxa11a?/?; hoxa11b?/?; hoxd11a?/? mutants have normal fin patterning. (D) waslbreph/+; hoxa11a?/?; hoxa11b?/?; mutants fail to form intermediate radials (bracket; n = 8, 88%). (E) hoxa13a?/?; hoxa13b?/? mutants fail to form distal fin rays (asterisk). (F) waslbreph/+; hoxa13a?/?; hoxa13b?/?; hoxd13a+/? mutants show enhanced PD elaboration with multiple intermediate radials (arrowheads). (G) Representative confocal images of hoxa11b:tdTom expression in early pectoral fins. (H) Boxplot of pixel intensity reveals higher hoxa11b:tdTom expression in mutant fins. Intensity is significantly elevated in reph homozygotes at 6 dpf (double asterisk, n = 5 each genotype, Welch?s Two Sample t test, t = 5.51, p = 0.0015) and in reph heterozygotes at 9 dpf (single asterisk, n = 4 each genotype, Welch?s Two Sample t test, t = 2.45, p = 0.035). See also Figures S5, S6, and S7 EXPRESSION / LABELING:
|
|
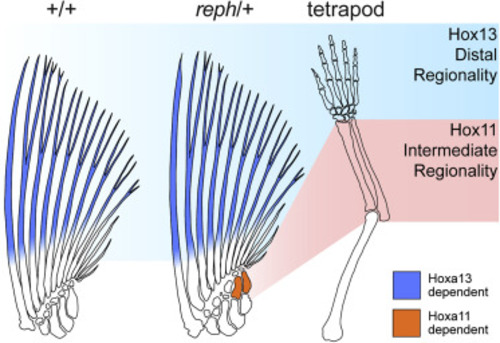
Figure 7. Intermediate radials share a developmental identity with the limb zeugopod Outlines of skeletons of wild-type and reph/+ mutant zebrafish pectoral fins and tetrapod limb. Both fins and limbs exhibit regional dependence on Hox cognate groups to properly form bones. Distal regions are dependent on Hox13 genes for their formation (blue background shading). Dermal fin rays (blue) are severely reduced in the absence of Hoxa13 paralogs, while the limb autopod does not form in the absence of Hoxa13 and Hoxd13. "Intermediate" regions are dependent on Hox11 genes (red background shading). The limb zeugopod fails to form long bones with loss of Hoxa11 and Hoxd11. Similarly, in reph/+ mutants, intermediate radials (orange) are not formed when Hoxa11 paralogs are lost. An intermediate regional dependence is not present in wild-type fins. |
|
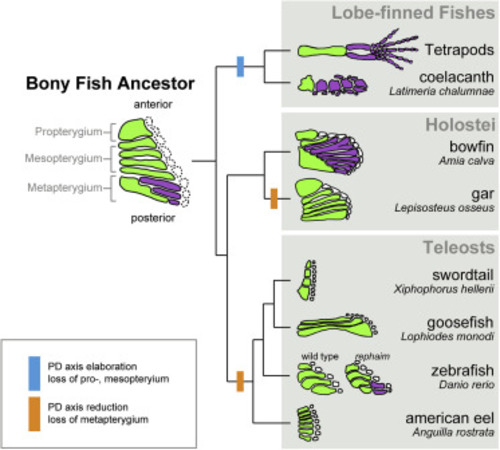
Figure S1. Phylogenetic patterns of pectoral appendage articulation in bony fishes, related to Figure 1 The last common ancestor of the bony fishes (the clade that encompasses the teleost and tetrapod lineages) had a polybasal condition in which the most proximal endochondral elements of the fin articulated with the shoulder girdle. The pectoral fin consisted of the anterior propterygium, the middle mesopterygium, and the posterior metapterygium. In the lineage leading to tetrapods, the propterygium and mesopterygium are postulated to have been lost along with the dermal fin rays, while new long bones that articulate end on end were added along the PD axis of the metapterygium, ultimately forming the limb. Conversely, in the teleost lineage, it is proposed that the metapterygium was lost while the propterygium and mesopterygium were retained, and no teleost exhibits end-on-end articulation of long bones in the pectoral fin. While rare, variation in the teleost pectoral fin endoskeleton is observed in proximal radial length as well as the number of radials along the AP axis of the fin in different clades. However, no teleost species showing PD long bone articulation comparable to that seen in tetrapods have been described. |
|
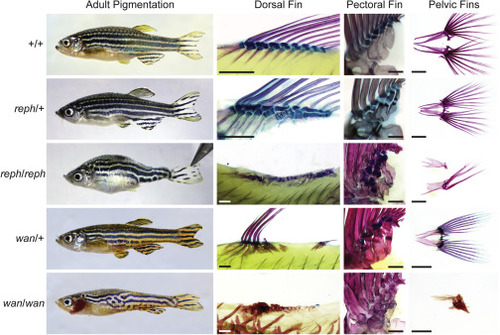
Figure S2. Additional adult phenotypes of reph and wan mutants, related to Figure 1 Adult pigmentation and skeletal patterning phenotypes of dorsal, pectoral, and pelvic fins in reph and wan mutants and wild-type adults. reph heterozygotes show increased number of radials in the dorsal fin (asterisk), however pelvic fins are unaffected. reph homozygous mutants show more severe effect on pigmentation and defects in lepidotrichia growth and maintenance. Dorsal and pectoral fin endoskeleton in homozygous fish is uninterpretable. wan heterozygotes show alteration in pigmentation and medial fin growth and patterning, while pelvic fins are unaffected. Similar to reph, wan homozygous fish show a more severe effect on pigmentation, and have highly dysmorphic fin rays and endoskeletal elements in the dorsal, pectoral, and pelvic fins. Scale bars 500 ?m in dorsal fin panels, 2 mm in pectoral fin panels, and 1 mm in pelvic fin panels. |
|
Figure S3. Mutations in waslb and vav2 cause the reph and wan phenotypes, respectively, related to Figure 1 (A) Mapping score from homogeneity-by-descent analysis for reph and wan mutants based on whole-exome sequencing of homozygous populations. Asterisks highlight regions of putative linkage. (B) Fine mapping of the chromosome with the highest mapping score by recombination analysis in individual homozygous mutants. Shown are informative markers used, their position, and the number of recombinants identified per meiosis scored. Within the smaller linked interval, sequence data from the mutant pool was used to identify nonsynonymous SNPs as candidate mutations responsible for the altered skeletal phenotype of reph and wan. (C, D) Protein schematics of Waslb (C) and Vav2 (D) showing positions of missense mutations and CRISPR-generated lesions. Multiple sequence alignment reveals conservation of the mutated residues across vertebrates. Mutant alleles are listed with their consequence on amino acid sequence and their phenotypic effect. (E-G) HeLa cells transfected with constructs encoding N-terminal GFP fusions to mouse wild-type WASL (GFP-WASL, E) and an WASL version containing the orthologous reph mutation S249P (GFP-WASL, F). (G) Qualitative scoring of transfected cells; white, predominantly cytoplasmic localization; black, predominantly nuclear localization; gray, no bias observed in localization. (H) Summary of known protein-protein interactions that link Wasl and Vav2 proteins. Vav2 acts as a guanine nucleotide exchange factor for Cdc42, converting Cdc42-GDP to Cdc42-GTP. GTP-bound Cdc42 is a regulator of Wasl activity. (I) Phenotype of cis frameshift and cis in-frame lesions upstream of the reph mutation. Scale bars 2 mm. |
|
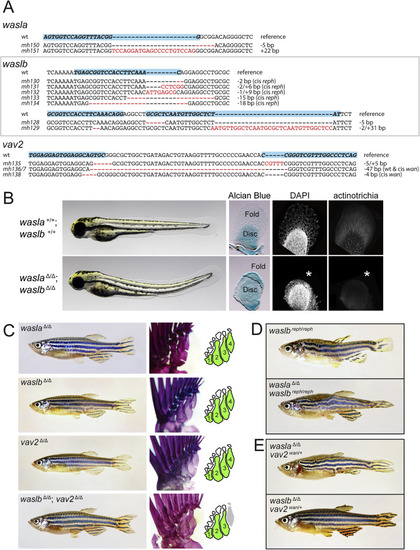
Figure S4. CRISPR-generated loss-of-function mutants of waslb and vav2 and their interaction with reph and wan, related to Figure 1 (A) Mutant alleles of wasla, waslb, and vav2 generated by CRISPR-Cas9 targeted gene editing. Shown is reference wild-type sequence as well as the sequence for the generated mutant alleles. Guide target positions are indicated in bold type surrounded by a blue box. (B) wasla; waslb double mutants at 3 dpf. Double mutants exhibit curvature of the body and severe reduction of the fin fold. Alcian Blue staining shows that the endoskeletal disc is largely unaffected. Immunolabelling of collagen reveals that the smaller fin fold is devoid of actinotrichia (asterisk). (C) Adult phenotypes of wasla, waslb, and vav2 mutants. Homozygous single mutants of each gene are phenotypically wild type and exhibit typical fin patterning (n = 10 for each mutant). waslb; vav2 double mutants exhibit loss of proximal radial 4 (n = 4). (D) Loss of wasla function does not affect expression of the reph phenotype. (E) Loss of wasla function does not affect expression of the wan phenotype, while loss of waslb rescues the wild-type phenotype in wan/+ mutants. |
|
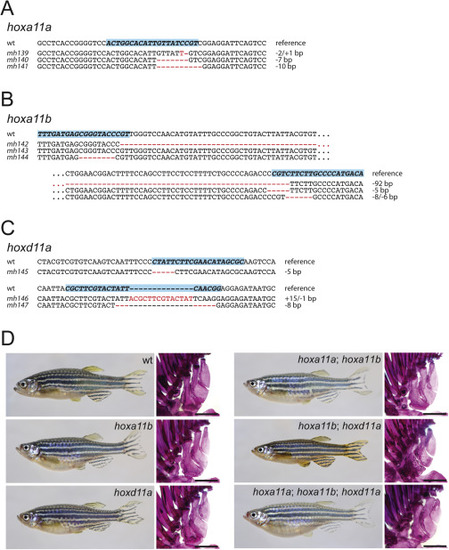
Figure S5. CRISPR-generated hox11 loss-of-function mutants, related to Figure 6 Mutant lines of hox11 paralogs. (A) hoxa11a (B) hoxa11b (C) hoxd11a generated by CRISPR-Cas9 targeted gene editing. Shown is the targeted wild-type sequence as well as the sequence for the generated mutant alleles (blue box indicates guide target sequence). (D) Representative singular and compound mutants. All mutant combinations are viable and fertile. Scale bars 500 ?m. |
|
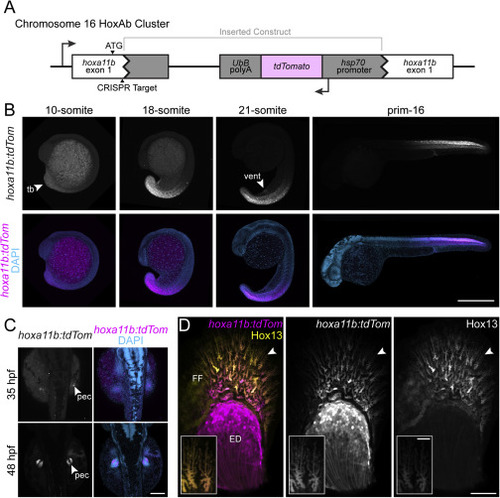
Figure S6. Targeted reporter construct knock-in recapitulates endogenous hoxa11b expression patterns, related to Figure 6 (A) Schematic of hoxa11bTg(hsp70:tdTomato) (hoxa11b:tdTom) locus. The transgene is inserted in the reverse orientation relative to the endogenous promoter. (B) Early axial expression in the tailbud, flank, and spinal cord in lateral view. (C) Expression in early fin buds in dorsal view. (D) At 3 dpf, fin fold cells are labeled by both hoxa11bTg(hsp70:tdTomato) and the Hox13 reporter Tg(2P?epi:eGFP), while only hoxa11bTg(hsp70:tdTomato) marks the endoskeletal disc. Anterior to top in C, distal to top and anterior to left in D; ED, endoskeletal disc; FF, fin fold; pec, pectoral fin; tb, tail bud; scale bars (B) 500 ?m, (C) 125 ?m, (D) 100 ?m (10 ?m inset). |
|
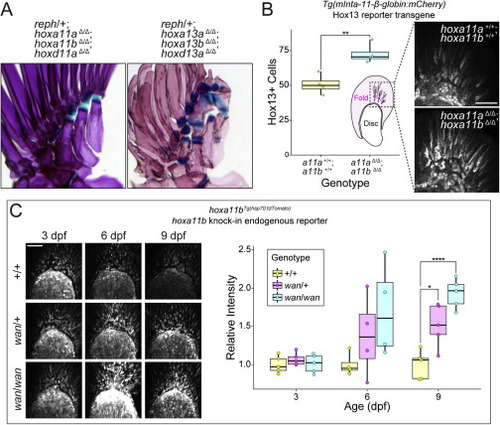
Figure S7. Interactions between Hox genes and reph and wan mutations, related to Figure 6 (A) Genetic interactions between reph and posterior Hox genes shape expression of the intermediate radial phenotype. reph heterozygotes that are triply homozygous null for hoxa11a, hoxa11b, and hoxd11a develop wild-type pectoral fin endoskeletons without intermediate radials. Heterozygous reph mutants triply homozygous for hoxa13a, hoxa13b, and hoxd13a exhibit an enhance endoskeleton elaboration phenotype, with each proximal radial producing intermediate radials. (B) HoxA11 paralogs negatively regulate Hox13 function in the fin. Double null mutants of hoxa11a and hoxa11b have significantly more Hox13-positive cells in the fin fold than do wild-type animals at 3 dpf (n = 4 each genotype, Welch?s Two Sample t test, t = 4.42, p = 0.0044). (C) hoxa11b expression is increased in wan mutants. Similar to reph mutants, wan mutants show higher levels of hoxa11b:tdTom expression in the early fin bud after 3 dpf. Intensity is significantly elevated in wan heterozygotes (single asterisk) and homozygotes (quadruple asterisk) at 9 dpf (n = 5 each genotype, Welch?s Two Sample t test, heterozygotes t = 3.43, p = 0.0055; homozygotes t = 7.82 p = 0.000026). Anterior to left, distal to top in all fin panels. Scale bars (B, C) 50 ?m. |
Reprinted from Cell, 184(4), Hawkins, M.B., Henke, K., Harris, M.P., Latent developmental potential to form limb-like skeletal structures in zebrafish, 899-911.e13, Copyright (2021) with permission from Elsevier. Full text @ Cell