- Title
-
The DNA Damage-Regulated Autophagy Modulator DRAM1 Links Mycobacterial Recognition via TLR-MYD88 to Autophagic Defense
- Authors
- van der Vaart, M., Korbee, C.J., Lamers, G.E., Tengeler, A.C., Hosseini, R., Haks, M.C., Ottenhoff, T.H., Spaink, H.P., Meijer, A.H.
- Source
- Full text @ Cell Host Microbe
|
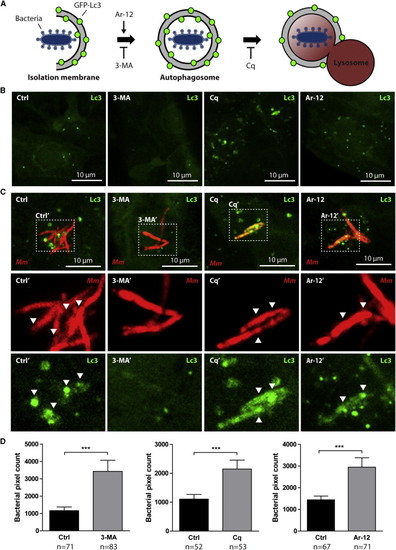
Stress-Induced Autophagy Is Not Beneficial for Defense of Zebrafish Embryos against Mycobacterial Infection (A) Schematic representation of the effects of 3-MA, Cq, and Ar-12 on autophagosome formation and autophagic flux. (B) GFP-Lc3 embryos 2 dpf were treated for 24 hr with DMSO (control), 3-MA, Cq, or Ar-12. Representative confocal micrographs of endothelial cells at 3 dpf are shown. (C) GFP-Lc3 embryos treated as described for (A) injected with Mm. Representative confocal micrographs of infected cells at 3 dpf are shown. Boxed areas are detailed below; arrowheads indicate overlap between Mm and Lc3. (D) AB/Tupfel long fin (AB/TL) embryos treated as described for (A) infected with Mm. Bacterial pixel counts were determined at 3 dpi. Data (mean ± SEM) is pooled from at least two individual experiments (n e 50 embryos per group). See also Figure S1. EXPRESSION / LABELING:
|
|
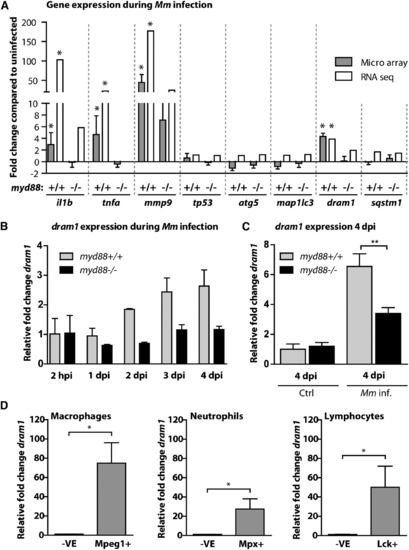
dram1 Is Induced by Mycobacterial Infection in a Myd88-Dependent Fashion (A) myd88+/+ and myd88-/- embryos infected with Mm were snap frozen individually at 4 dpi, and triplicate samples were compared with PBS-injected controls using a common reference microarray design. Observed differences were confirmed by RNA sequencing of pools (n = 20 embryos) of uninfected and Mm-infected myd88+/+ and myd88-/- embryos. (B) Expression of dram1 at multiple time points after infection was analyzed by qPCR on pools of Mm-infected myd88+/+ and myd88-/- embryos, relative to PBS-injected controls (mean ± SEM of n = 2 biological replicates with 20 embryos per pool). (C) Expression levels of dram1 at 4 dpi in individual myd88+/+ and myd88-/- embryos with or without infection were determined by qPCR (mean ± SEM of n = 3 biological replicates with 10 embryos per pool). (D) Macrophages, neutrophils, and leukocytes from 5-6 dpf larvae were isolated by FACS. Expression of dram1 in the positive fractions (e.g., Mpeg1+) relative to the negative fractions (-VE) was determined by qPCR (mean ± SEM of n = 4 biological replicates). See also Figure S2. |
|
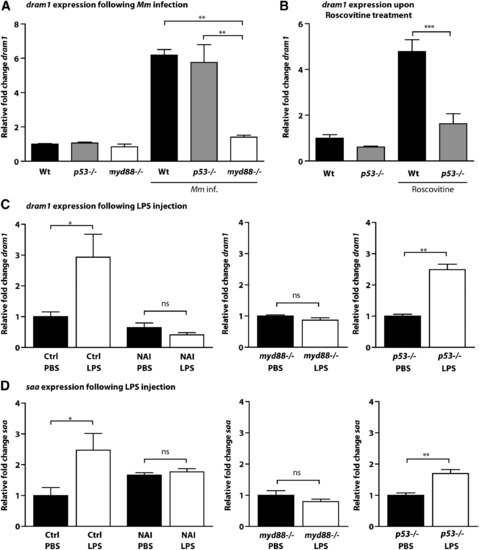
dram1 Expression during Mycobacterial Infection Is Independent of p53 but Dependent on NF-κB (A-C) Expression levels of dram1 were determined by qPCR for (A) wild-type, p53-/-, and myd88-/- embryos 4 days after infection with Mm, relative to mock-injected controls; (B) wild-type and p53-/- embryos at 5 dpf after 24 hr of treatment with roscovitine, relative to untreated controls (see also Figure S3); and (C) 1 dpf embryos at 2 hpi with LPS, relative to their respective PBS-injected controls (left panel: wild-type embryos with or without NAI treatment [4 hr total, including 2 hr pretreatment]; middle panel: myd88-/- embryos; right panel: p53-/- embryos). (D) Expression levels of saa were determined by qPCR under the same conditions as those described for (C). All graphs show data (mean ± SEM) from three biological replicates with n = 20 embryos pooled per replicate. See also Figures S2 and S3. |
|
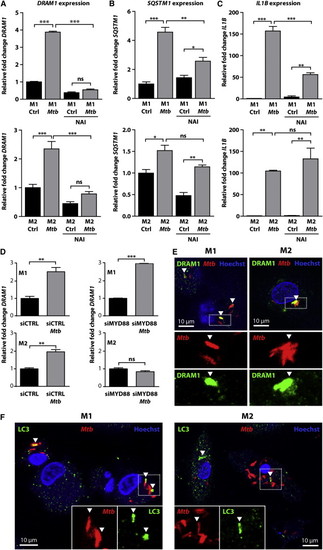
DRAM1 Colocalizes with Mtb and Is Regulated by MYD88-NF-κB Signaling in Human Primary Macrophages (A?C) The effect of NAI treatment on expression of (A) DRAM1, (B) SQSTM1, and (C) IL1B in M1 and M2 in the presence or absence of Mtb was determined by qPCR, relative to uninfected controls. All graphs show data (mean ± SEM) from three biological replicates. (D) Expression of DRAM1 in M1 and M2 transfected with siCTRL or siMYD88 with and without Mtb infection. All graphs show data (mean ± SEM) from three biological replicates. (E and F) Immunohistochemistry for (E) DRAM1 (green) or (F) LC3 (green) performed on M1 and M2 at 72 hpi with Mtb (red); Hoechst staining (blue) was used to outline the nuclei and cell boundaries. See also Figure S4. |
|
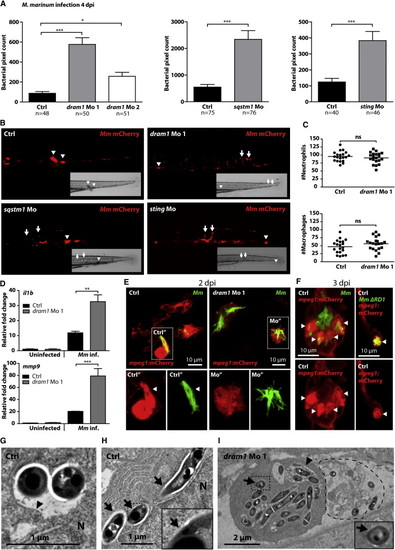
Dram1 Is Required to Contain Mm Growth inside Macrophages (A) Zebrafish embryos injected with standard control morpholino, morpholino against dram1 (dram1 Mo 1 and Mo 2), sqstm1 (sqstm1 Mo), or sting (sting Mo) were injected with mCherry-labeled Mm (see also Figure S5). Bacterial pixel counts were determined at 4 dpi. Data (mean ± SEM) is pooled from at least two individual experiments (n = 48?76 embryos per group). (B) Stereo micrographs of the tail region of highly infected standard control, dram1 Mo 1-, sqstm1 Mo-, or sting Mo-injected embryos. Arrowheads indicate granuloma formation, and arrows indicate accumulation of bacteria in intersegmental veins. (C) Total numbers of neutrophils and macrophages in 2 dpf control- or dram1 Mo 1-injected embryos were quantified using a fluorescence stereo microscope (n = 20 per condition, blinded). Total numbers of macrophages were determined by performing whole-mount L-plastin immunohistochemistry and deducting the number of cells positive for neutrophil-specific myeloperoxidase activity from the number of L-plastin-positive total leukocytes per embryo. Each data point represents an individual embryo, and lines indicate the mean. (D) Expression levels of il1b and mmp9 at 4 dpi in control morpholino-injected and dram1 Mo 1-injected embryos with or without Mm infection (200 colony-forming units [cfu] for control and 50 cfu for dram1 Mo 1 to obtain equal bacterial burdens) were determined by qPCR (mean ± SEM from n = 3 biological replicates with 20 embryos per pool). (E and F) Representative confocal micrographs of mpeg1:mCherry transgenic embryos injected with control or dram1 morpholino 1 and infected with GFP-labeled Mm or ΔRD1 Mm (E) 2 dpi or (F) 3 dpi. Boxed areas are detailed below, with the green and red channels shown separately. Arrowheads indicate bacteria enclosed by membranes. (G?I) Transmission electron micrograph of control morpholino-injected embryos with bacteria inside a phagosome (G) or bacteria in the cytoplasm (H) or dram1 morpholino-injected embryos with bacteria present both in phagosomes and in the cytoplasm (I). Arrows indicate cytoplasmic bacteria, while arrowheads indicate phagosomal membranes. Boxed areas are enlarged in the insets, and the dashed line in (I) indicates the remnants of a dead infected cell. See also Figure S5. |
|
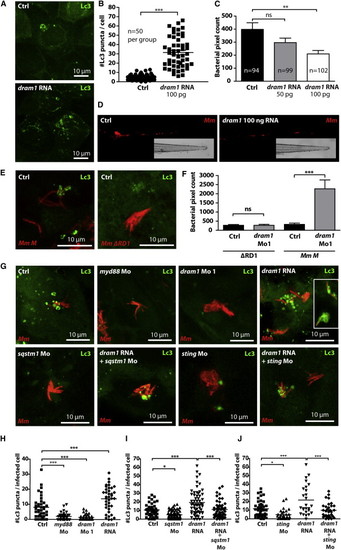
Dram1 Modulates an Autophagic Defense Mechanism that Requires Bacterial RD1 Virulence, Host p62/Sqstm1, and Sting (A) GFP-Lc3 embryos were injected with 100 pg dram1 RNA. Representative confocal micrographs of epithelial cells at 3 dpf are shown. (B) The number of GFP-Lc3 punctae was determined for n = 50 cells per group and quantified based on confocal micrographs of control- and dram1 RNA-injected embryos (n e 5, blinded). (C) Bacterial pixel counts were determined at 3 dpi for infected control- or dram1 RNA-injected embryos. Data (mean ± SEM) is pooled from two individual experiments (n e 94 embryos per group). (D) Representative stereo micrographs of the tail of infected control- or dram1 RNA-injected embryos at 3 dpi. (E) Representative confocal micrographs of GFP-Lc3 embryos infected with Mm M ΔRD1 (400 cfu) and wild-type Mm M bacteria (200 cfu). (F) Bacterial pixel counts following these infections were determined with or without dram1 knockdown. Data (mean ± SEM) are pooled from two individual experiments (n > 62 embryos per group). (G) Representative confocal micrographs of GFP-Lc3 embryos injected with control morpholino, myd88 morpholino, dram1 morpholino 1, dram1 RNA (100 pg; inset in the image is a micrograph from a different RNA-injected embryo), sqstm1 morpholino, dram1 RNA + sqstm1 morpholino, sting morpholino, and dram1 RNA + sting morpholino. All groups were injected with Mm. (H?J) Confocal micrographs were used to quantify GFP-Lc3 punctae per infected cell (n e 5 embryos per group, blinded) to evaluate the effect of dram1 RNA (H), dram1 RNA combined with sqstm1 Mo (I), or dram1 RNA combined with sting Mo (J). See also Figure S6. PHENOTYPE:
|
|
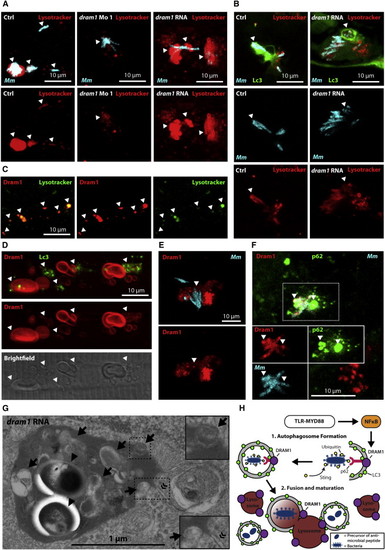
Dram1 Mediates Autophagic Flux and Lysosomal Maturation via Multiple Vesicle Fusion Events (A and B) Embryos were infected with crimson-labeled Mm and stained with LysoTracker Red (arrowheads indicate colocalization). (A) Wild-type embryos injected with standard control, dram1 morpholino, or dram1 RNA (100 pg). (B) dram1 RNA- or control-injected GFP-Lc3 embryos. (C?F) Embryos transiently expressing mCherry-Dram1, colocalized with (C) LysoTracker Green, (D) GFP-Lc3, (E) crimson-labeled Mm, or (F) crimson-labeled Mm, and immunohistochemistry detection of p62 (arrowheads indicate colocalization). (G) Transmission electron micrograph of dram1 RNA-injected embryos infected with Mm (arrowheads). Arrows indicate (remnants of) vesicle fusion, and « indicates the double membrane of an autophagosome. (H) Schematic representation of the findings presented in this manuscript, as explained in the Discussion. See also Figure S7. PHENOTYPE:
|
|
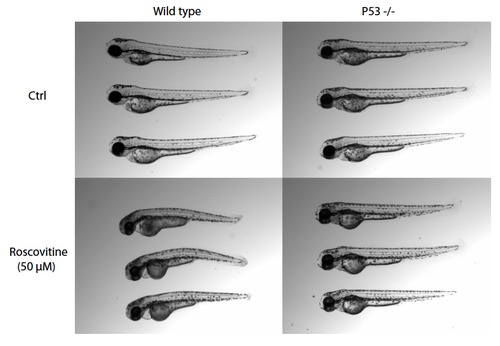
p53-/- embryos are resistant to roscovitin-induced cell death. Representative images are shown for AB/TL (wild type) and p53-/- embryos at 5 days post fertilization after 24 hours of treatment with 50 M roscovitine (treatment started at 2 dpf ). |
|
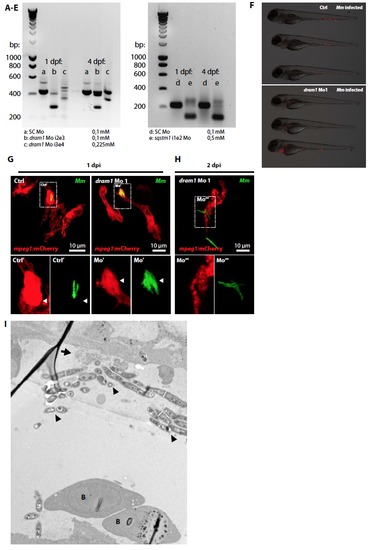
Dram1 knockdown effect in zebrafish embryos. Reverse transcription polymerase chain reaction (RT-PCR) was used to confirm antisense morpholino blocking of intron-exon splicing events in zebrafish dram1 or sqstm1 at 1 and 4 days post fertilization. (A) and (D) 0,1 mM Standard control (SC) morpholino; (B) 0,1 mM dram1 Mo 1 (i2e3, targeting the splicing event between the second intron and the third exon); (C) 0,225 mM dram1 Mo 2 (i3e4, targeting the splicing event between the third intron and the fourth exon); (E) 0,5 mM sqstm1 Mo (i1e2, targeting the splicing event between the second intron and the third exon). For both dram1 and sqstm1, the RT-PCR product (approximately 400 and 200 bp in size, respectively) is disrupted upon morpholino injection. Morpholino sequences and primers used for the RT-PCR are listed in supplementary tables 1 and 2. Supplementary figure 5, related to figure 5. Dram1 knockdown effect in zebrafish embryos (F) figure 5. Equal bacterial burdens in control and dram1 morpholino injected embryos. Representative stereo micrographs of standard control or dram1 morpholino injected embryos. Control injected embryos were infected with 200 CFU Mm versus 50 CFU for dram1 morphants to obtain equal bacterial burdens at the time point of RNA isolation. (G-H) Mycobacterial localization in or outside of macrophages. Representative confocal micrographs of mpeg1:mCherry transgenic embryos injected with standard control or dram1 morpholino 1 and infected with GFP-labeled Mm (A: 1 dpi; B: 2 dpi). Boxed areas are detailed below with the green and red channel shown separately. Arrowheads indicate bacteria enclosed by membranes. (I) Transmission electron micrograph of a dram1 morpholino injected embryo infected with Mm. Clusters of extracellular bacteria (arrowheads) are residing inside a blood vessel, as illustrated by the presence of red blood cells (B). The remnants of a ruptured cell are indicated by an arrow. |
|
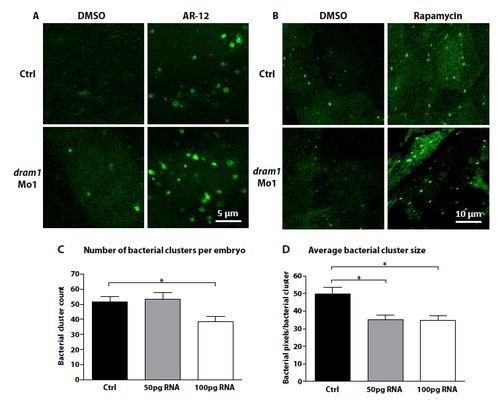
Dram1 RNA and defense against mycobacterial infection. (A-B) The autophagy response initiated by Dram1 is distinct from that initiated by Ar-12 or rapamycin treatment. Representative confocal micrograph of standard control or dram1 morpholino injected embryos treated with (A) Ar-12, (B) rapamycin, or DMSO as a control. Embryos were treated at 2dpf for 24 hours. (C-D) Dram1 overexpression effect on the formation of granuloma-like aggregates. Wild type embryos were mockinjected or injected with 50 or 100 pg dram1 RNA immediately after fertilization. Embryos were infected with 200 CFU Mm at 1 dpf. Whole embryo stereo fluorescent micrographs were taken at 3 dpi and (A) the total number of bacterial clusters and (B) the average bacterial cluster size were determined using dedicated software (Stoop et al., 2010). Data (mean ± SEM) is pooled from two individual experiments (n>94 embryos per group). Significant differences were calculated by one-way ANOVA with Tukey?s Multiple Comparison method as a post-hoc test (*= p<0.05). Significant reduction of both the total number of clusters and cluster size by dram1 overexpression is in agreement with the reduction of total bacterial pixel counts shown in figure 6C. |
|
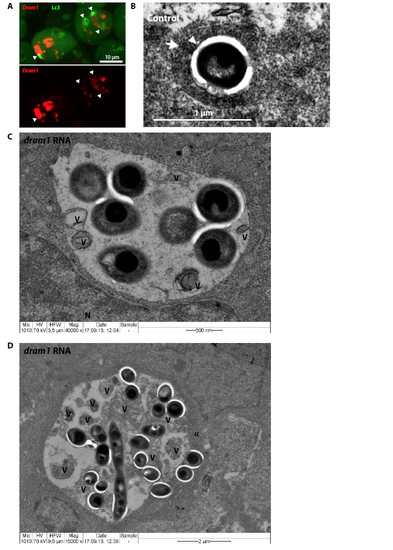
Effect of Dram1 on autophagosomal vesicles and fusion events. (A) Representative confocal micrograph of GFP-Lc3 embryos transiently expressing mCherry- Dram1. Co-localization is indicated with arrowheads. (B-D) Transmission electron micrograph of control or dram1 RNA injected embryos infected with Mm. (B) Bacteria (indicated by an arrowhead) inside an autophagolysome, as characterized by the presence of cytoplasmic material inside a singlemembraned vesicle (indicated by an arrow). (C-D) Two examples of large bacteria-containing vesicles, as frequently observed in dram1 overexpressed embryos infected with Mm. V= remnants of vesicular fusion; N = nucleus; « = doublemembrane. |