- Title
-
Fine-tuning of Hh signaling by the RNA-binding protein Quaking to control muscle development
- Authors
- Lobbardi, R., Lambert, G., Zhao, J., Geisler, R., Kim, H.R., and Rosa, F.M.
- Source
- Full text @ Development
|
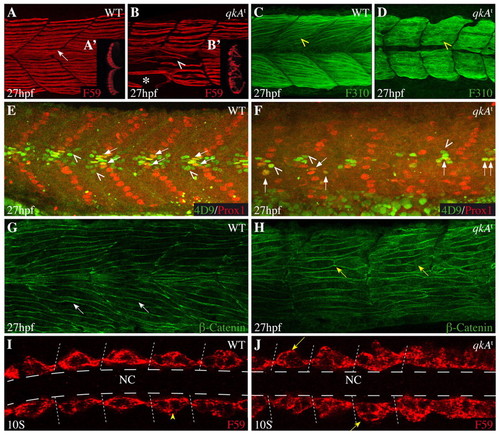
Requirement for qkA during somite development. (A-H) Lateral views from 27-hpf zebrafish embryos of the indicated genotypes (upper right) immunostained with the indicated markers (lower right) and analyzed by confocal imaging. Anterior is to the left. (A-B2) Slow muscle cells labeled by F59 (arrow) are disorganized in qkAt homozygotes (arrowhead), are less numerous, exhibit gaps (asterisk) and invade the fast muscle domain (see optical transverse sections in insets A2,B2). (C,D) Fast muscle fibers (yellow arrowhead) appear immature in qkAt homozygotes, with predominantly cytosolic staining and imprecise contours. (E,F) Muscle subpopulations are reduced in qkAt. Embryos were stained for Prox1 [slow muscle cells, including muscle pioneers (MPs)] and Engrailed [MPs (arrows) and medial fast fibers (MFFs; arrowheads) by 4D9 antibody]. Note the reduction in all these cell types in the qkAt homozygotes. (G,H) qkA is required for fast myotube formation. β-catenin staining reveals a large number of short mononucleated cells (yellow versus white arrows) in the fast muscle domain, indicating retarded fusion. (I,J) Early requirement for qkA in slow muscle development. Ten-somite stage embryos stained for slow muscle, dorsal view. At this stage, adaxial cells abut the notochord in wild type (WT) (arrowhead), whereas mutant somites often exhibit several mislocated cells (arrows). Dashed lines indicate the position of the notochord. NC, notochord. EXPRESSION / LABELING:
PHENOTYPE:
|
|
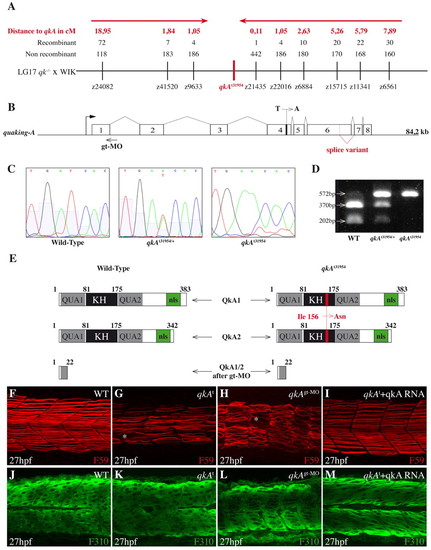
Molecular and functional characterization of the qkA locus. (A) Genetic mapping of zebrafish quaking A (qkA). On chromosome 17, the qkAt31954 mutation is linked to markers z9633 and z21435, delineating a candidate genomic region of ~100 kb that contains qkA. Red arrows indicate the directions of mapping progression. (B) qkA gene structure. qkA includes eight exons. The alternative splicing site and the positions of the qkAt31954 mutation in exon 4 and of the qkAgt-MO are indicated. (C) Chromatogram showing the single base change in qkAt31954 mutants. (D) qkAt31954 mutation leads to the loss of an RFLP site (BclI). The qkA region was PCR amplified from genomic DNA and digested with BclI, which normally cuts a 572 bp fragment into 370 bp and 202 bp fragments. (E) Structure of predicted QkA protein isoforms in WT, mutant and qkAgt-MO embryos. The t31954 mutation leads to the Ile156Asn amino acid change (red) in the KH domain. KH, K homology domain; nls, nuclear localization signal. (F-M) qkAt31954 mutation can be rescued by qkA mRNA injection and is phenocopied by qkAgt-MO. WT, qkAt31954 mutants or qkA morphants, untreated or injected with qkA mRNA, were fixed at 27 hpf and processed as in Fig. 1A-D. Asterisks, gaps in slow muscle cells. |
|
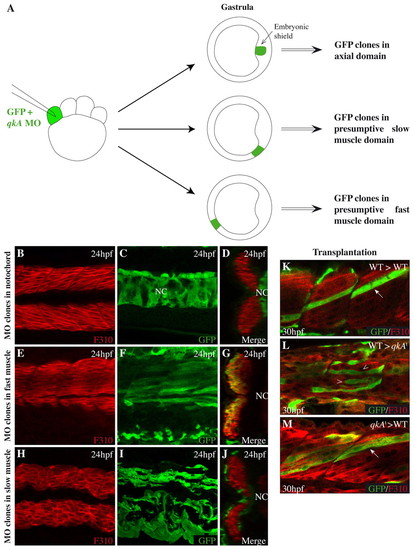
QkA activity is not required in fast muscle cells. (A) Scheme for the generation of morphant clones. qkAgt-MO was injected together with GFP RNA at the 8-cell stage and embryos were sorted for the dorsoventral position of the clone at the onset of gastrulation. (B-J) Embryos were immunostained at 24 hpf for fast muscle (F310) and GFP and analyzed by confocal microscopy. (B,C,E,F,H,I) Lateral views, anterior to the left; (D,G,J) corresponding optical sections (merge). (K-M) GFP-labeled WT (K,L) or mutant (M) cells were transplanted into WT (arrows; K,M) or qkAt31954 homozygotes (arrowheads; L) and analyzed for fast muscle development at 30 hpf. NC, notochord. |
|
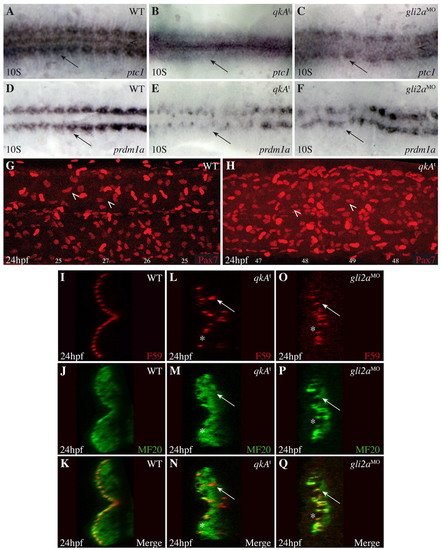
Convergence of qkA and Hh phenotypes. (A-F) qkAt and gli2aMO zebrafish embryos exhibit downregulated Hh signaling. Embryos were processed at the 10-somite stage for in situ hybridization with the indicated probes. Dorsal view. (G,H) Lateral views of Pax7-immunostained embryos at 24 hpf. Two populations of Pax7-positive cells can be discerned, of which dermomyotomal cells (arrowheads) exhibit the weaker expression. The number of dermomyotomal cells per somite is indicated beneath each somite. (I-Q) Slow fiber distal migration requires QkA and Hh activities. Embryos were stained at 24-hpf for slow fibers (F59) and all differentiated muscle fibers (MF20) as indicated, then analyzed by confocal microscopy (transverse sections). Note the presence of gaps (asterisks) and deep slow muscle cells (arrows) medial to the fast muscle cells in qkAt mutants and gli2a morphants. EXPRESSION / LABELING:
PHENOTYPE:
|
|
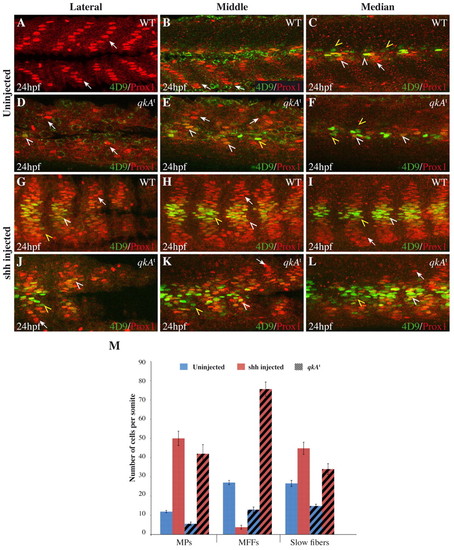
Requirement for qkA in Hh signaling. (A-L) WT or qkAt mutant zebrafish embryos were injected with shh RNA (G-L) or left untreated (A-F), then fixed at 24 hpf and stained for Prox1 and Engrailed (4D9) as in Fig. 1 revealing slow fibers (arrows), MPs (white arrowheads) and MFFs (yellow arrowheads). Different mediodistal sections are shown, as indicated above each column. (M) The numbers of each fiber type per somite (based on the expression of Prox1 and/or Eng) plotted according to the genotype and the treatment (n=5-6 embryos per condition; see Table 1). Error bars indicate s.d. |
|
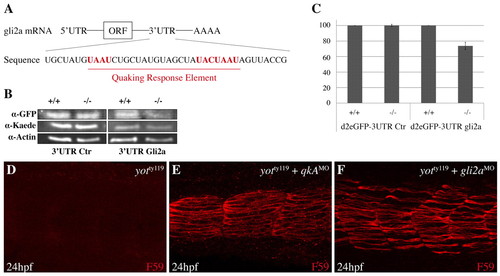
Analysis of QkA requirement for gli2a mRNA stability. (A) The region of the zebrafish gli2a 32UTR mRNA that contains the Quaking response element (QRE). Consensus sequences are in red. (B,C) Biosensor assay for QkA activity. RNA encoding d2eGFP flanked by the gli2a 32UTR or a control (Ctr) 32UTR were injected, together with kaede RNA as a control, into WT (+/+) or qkAt mutants (-/-). (B) Proteins from each embryo were extracted at the 10-somite stage and probed by western blot. (C) Protein expression values from B were normalized to controls (see Materials and methods) and are plotted according to the genotype. Error bars indicate s.d. (D-F) Slow muscle fiber staining in yotty119 embryos injected with qkAATG-MO or gli2aMO. |
|
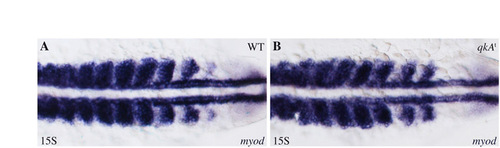
myod is not perturbed in qkAt31954. (A,B) Whole-mount in situ hybridization using a myod probe at the 15-somite stage. In the qkAt mutant (B), the myod MRF gene exhibits an expression profile that is similar to that of WT embryos (A), showing that Myogenesis regulatory factor (MRF) gene expression is not responsible for the muscle phenotype. Dorsal views, anterior to the left. |
|
The qkAgt-MO induces an alternative splicing site. (A) RT-PCR on uninjected or qkAgt-MO-injected embryos. Two bands corresponding to the two qkA isoforms are amplified. A shift of both isoforms is seen in morphants. (B) Sequencing of each PCR-amplified isoform after gel extraction. With qkAgt-MO, a cryptic splicing site is revealed that allows a short part of intron 1 to be transcribed. This induces a frameshift with the appearance of a stop codon in the intron 1 sequence (blue) interrupting qkA1 or qkA2 mRNA translation. L, 1 kb DNA ladder: lower band, 1 kb; upper band, 1.5 kb. |
|
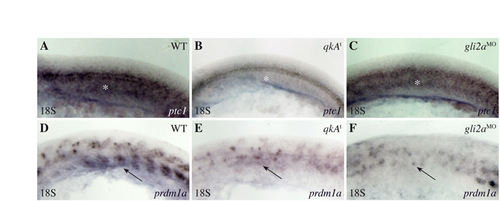
Reduction of the Hh signaling pathway at the 18-somite stage in qkAt and gli2aMO. (A-F) qkAt and gli2aMO embryos exhibit hallmarks (white asterisks and black arrows point to the adaxial cell derivatives labeled by ptc1 and prdm1a, respectively) of downregulated Hh signaling. Embryos were fixed at the 18-somite stage and processed for in situ hybridization with ptc1 (A-C) and prdm1a (D-F) probes. Lateral views, anterior to the left. |
|
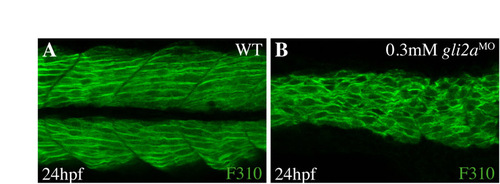
The loss of gli2a function affects fast muscle fiber development. (A,B) Confocal section of fast muscle fibers labeled by F310 in WT (A) and gli2a morphants (B) at 24 hpf. The fast muscle fibers lose their stereotyped diagonal orientation in the morphants. |
|
Partial restoration of the fast muscle cell phenotype at 48 hpf. (A-D) At 48 hpf, the slow muscle phenotype is not rescued in the mutant (B), which exhibits gaps (white asterisk) compared with WT embryos (A). The fast muscle fibers, constituting the second major muscle cell type, are organized in WT (C) in a stereotyped manner (diagonal direction of fibers, yellow arrowhead). At 48 hpf, in the qkAt mutant (D), the fast muscle fibers exhibit a myotube structure but are not as well organized as in the WT sibling (C, yellow arrowhead), indicating a partial rescue of the fast muscle fiber organization. Dorsal views, anterior to the left. |