- Title
-
Copper overload impairs hematopoietic stem and progenitor cell proliferation via prompting HSF1/SP1 aggregation and the subsequently downregulating FOXM1-Cytoskeleton axis
- Authors
- Li, L., Tai, Z., Liu, W., Luo, Y., Wu, Y., Lin, S., Liu, M., Gao, B., Liu, J.X.
- Source
- Full text @ iScience
|
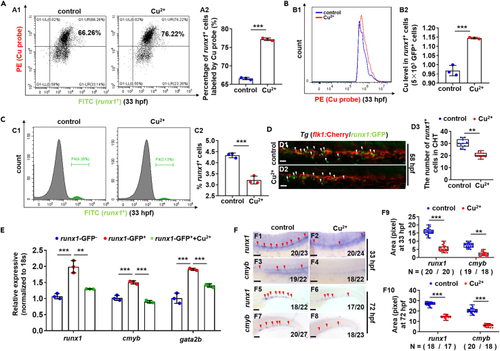
Accumulation of Cu in hematopoietic stem and progenitor cells (HSPCs) and the emergence and proliferation of HSPCs in Cu-stressed embryos (A and B) Proportion of HSPCs labeled with Cu probe (A) and the Cu content in individual runx1GFP+ cells (HSPCs) (B) were increased in Cu-stressed embryos at 33 hpf. A1, flow cytometry (FACS) plots; B1, flow cytometry (FACS) histogram; A2, calculation of the percentage of runx1GFP+ cells labeled by Cu probe; B2, calculation of Cu level in individual runx1GFP+ cells. (C) The percentage of runx1GFP+ cells in runx1:GFP embryos at 33 hpf. C1, flow cytometry (FACS) histogram; C2, calculation of the percentage of runx1GFP+ cells in different groups. (D) Cu-stressed flk1:Cherry/runx1:GFP embryos (D2) and the controls (D1) at 58 hpf. D3, calculation of runx1-positive cells (runx1+ cells). (E) The expression of HSPC genes runx1, cmyb, and gata2b in runx1GFP? cells, runx1GFP+ cells, and Cu-stressed runx1GFP+ cells, respectively. (F) The expression of HSPC genes runx1 and cmyb in embryos at 33 hpf and 72 hpf. F9, F10, calculation of runx1 and cmyb expression level in the Cu-stressed and control embryos, respectively. Each experiment was repeated three times, and a representative result is shown. Nchanged/Ntotal in the right bottom corner of each panel indicates embryos with changed expression/total tested embryos, and N in calculation panels indicates the number of embryos with changed expression in each group. The same for the numbers in the following figures. F1-F8, lateral view, anterior to the left, and dorsal to the up. Data are mean ± SD. t-test, ?p < 0.05, ??p < 0.01, ???p < 0.001. Scale bars, 20 ?m (D1-D2), 50 ?m (F1-F4), and 200 ?m (F5-F8). |
|
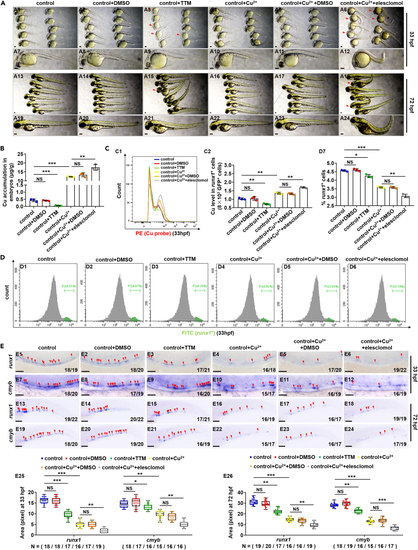
Cu chelator TTM and ionophore elesclomol affect embryonic and HSPC development during zebrafish embryogenesis (A) Phenotypes of zebrafish embryos in the control, DMSO exposed, TTM exposed, Cu exposed, Cu and DMSO co-exposed, or Cu and elesclomol co-exposed group, at 33 hpf and 72 hpf, respectively. (B) Cu concentration in zebrafish embryos from the control, DMSO exposed, TTM exposed, Cu exposed, Cu and DMSO co-exposed, or Cu and elesclomol co-exposed group at 33 hpf, respectively. (C) Cu content in individual runx1GFP+ cells (HSPCs) of zebrafish embryos in control, DMSO exposed, TTM exposed, Cu exposed, Cu and DMSO co-exposed, or Cu and elesclomol co-exposed group at 33 hpf, respectively. C1, flow cytometry (FACS) histogram; C2, calculation of Cu level in individual runx1GFP+ cells. (D) The percentage of runx1GFP+ cells in runx1:GFP embryos from different groups at 33 hpf, respectively. D1-D6, flow cytometry (FACS) histogram; D7, calculation of the percentage of runx1GFP+ cells in different groups. (E) The expression of HSPC genes runx1 and cmyb in embryos from different groups at 33 hpf and 72 hpf. E25, E26, calculation of runx1 and cmyb expression level in embryos from different groups, respectively. A1-A24, E1-E24, lateral view, anterior to the left, and dorsal to the up. Data are mean ± SD. t-test, ?p < 0.05, ??p < 0.01, ???p < 0.001. NS, not significant. Scale bars, 50 ?m (E1-E12), 100 ?m (A1-A24) and 200 ?m (E13-E24). |
|
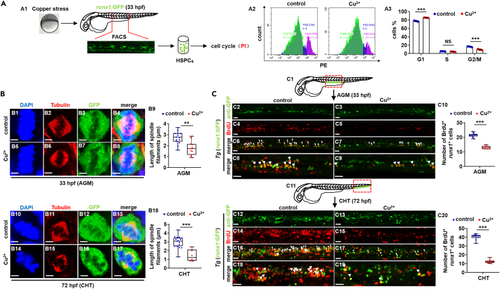
Cu overload induces HSPC proliferation impairment (A) Cell cycle of HSPCs in Cu-stressed embryos at 33 hpf. A1, Schema for the experiments; A2, flow cytometry (FACS) histogram; A3, cell cycle stage calculation. (B) Mitotic malformation of HSPCs in Cu-stressed embryos at 33 hpf (AGM) and 72 hpf (CHT), respectively. B1, B5, B10, B14, DAPI staining; B2, B6, B11, B15, anti-?-tubulin staining; B3, B7, B12, B16, anti-GFP staining; B4, B8, B13, B17, merged. At least 10 mitotic HSPCs in more than 10 embryos were observed for each group. B9, B18, calculation the length of spindles in metaphase runx1GFP+ cells at 33 hpf and 72 hpf, respectively. (C) BrdU cell proliferation assays in embryos at 33 hpf (C2-C9) and 72 hpf (C12-C19), respectively. C1, C11, AGM and CHT domain in embryos, respectively; C10, C20, calculation of the number of proliferative HSPCs in embryos from different groups. C2-C9, C12-C19, lateral view, anterior to the left, and dorsal to the up. Data are mean ± SD. t-test, ?p < 0.05, ??p < 0.01, ???p < 0.001. NS, not significant. Scale bars, 2 ?m (B1-B8, B10-B17) and 20 ?m (C2-C9, C12-C19). |
|
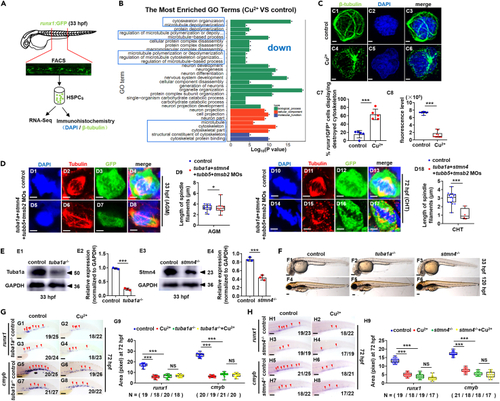
Cu overload destroys embryonic HSPC cytoskeleton (A) Schema for the experiments in (B) and (C). (B) Enriched GO terms for down-regulated DEGs in Cu-stressed HSPCs at 33 hpf. (C) Cytoskeleton protein immune-fluorescence for Cu-stressed HSPCs at 33 hpf. C1, C4, anti-?-tubulin staining; C2, C5, DAPI staining; C3, C6, merged; C7, the percentage of the Cu-stressed runx1GFP+ cells exhibiting destructive cytoskeleton; C8, calculation of the fluorescence intensity in each cell. (D) Mitotic malformation of HSPCs in the cytoskeleton gene morphants (tuba1a-MO, stmn4-MO, tubb5-MO, and tmsb2-MO together) at the 33 hpf (AGM) and 72 hpf (CHT), respectively. D1, D5, D10, D14, DAPI staining; D2, D6, D11, D15, anti-?-tubulin staining; D3, D7, D12, D16, anti-GFP staining; D4, D8, D13, D17, merged. At least 10 mitotic HSPCs in more than 10 embryos were observed for each group. D9, D18, calculation of the length of spindles in metaphase runx1GFP+ cells at 33 hpf and 72 hpf, respectively. (E) The protein level of Tuba1a (E1) and Stmn4 (E3) in tuba1a mutants and stmn4 mutants at 33 hpf, respectively. GAPDH was used as an internal control. (F) Phenotypes of tuba1a (F2, F5) mutants with a 4-bp deletion and stmn4 (F3, F6) mutants with an 8-bp deletion at 33 hpf and 120 hpf, respectively. (G) Expression of runx1 and cmyb in tuba1a mutants with or without Cu stresses at 72 hpf. G9, calculation of runx1 and cmyb expression in embryos from different groups. (H) Expression of runx1 and cmyb in stmn4 mutants with or without Cu stresses at 72 hpf. H9, calculation of runx1 and cmyb expression in embryos from different groups. F1-F6, G1-G8, H1-H8, lateral view, anterior to the left, and dorsal to the up. Data are mean ± SD. t-test, ?p < 0.05, ??p < 0.01, ???p < 0.001. NS, not significant. Scale bars, 1.2 ?m (C1-C6), 2 ?m (D1-D8, D10-D17), 100 ?m (D1-D3, F1-F6), and 200 ?m (G1-G8, H1-H8). |
|
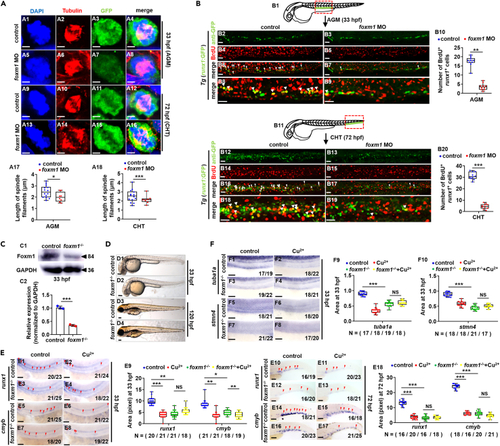
Dysfunction of gene foxm1 impairs the proliferation of HSPCs (A) Mitotic malformation of HSPCs in foxm1 morphants at 33 hpf (AGM) and 72 hpf (CHT), respectively. A1, A5, A9, A13, DAPI staining; A2, A6, A10, A14, anti-?-tubulin staining; A3, A7, A11, A15, anti-GFP staining; A4, A8, A12, A16, merged. At least 10 mitotic HSPCs in more than 10 embryos were observed for each group. A17, A18, calculation the length of spindles in metaphase runx1GFP+ cells at 33 hpf and 72 hpf, respectively. (B) BrdU cell proliferation in embryos at 33 hpf (B2-B9) and 72 hpf (B12-B19), respectively. B1, B11, AGM and CHT domain in embryos, respectively; B10, B20, calculation of HSPC proliferation in embryos from different groups. (C) The protein level of Foxm1 in foxm1 mutants with a 10-bp deletion at 33 hpf. GAPDH was used as an internal control. (D) Phenotypes of foxm1 mutants at 33 hpf and 120 hpf, respectively. (E) Expression of runx1 and cmyb in tuba1a mutants with or without Cu stresses at 33 hpf and 72 hpf. E9, E18, calculation of runx1 and cmyb expression in embryos from different groups. (F) Expression of tuba1a and stmn4 in foxm1 mutants with or without Cu stresses at 33 hpf. F9, F10, calculation of tuba1a and stmn4 expression in embryos from different groups. B2-B9, B12-B19, D1-D4, E1-E8, E10-E17, F1-F8, lateral view, anterior to the left, and dorsal to the up. Data are mean ± SD. t-test, ?p < 0.05, ??p < 0.01, ???p < 0.001, NS, not significant. Scale bars, 2 ?m (A1-A16), 20 ?m (B2-B9, B12-B19), 50 ?m (E1-E8, F1-F8), 100 ?m (D1-D4) and 200 ?m (E10-E17). |
|
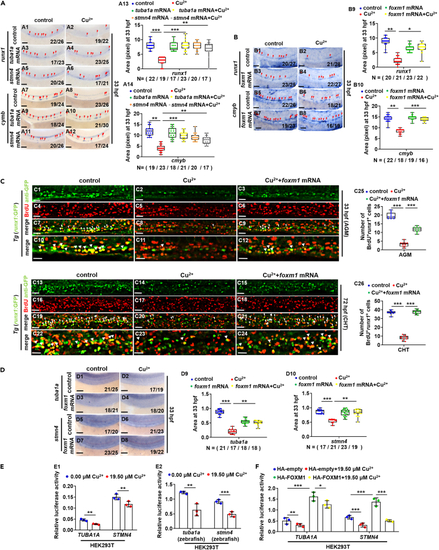
Ectopic expression of tuba1a, stmn4, or foxm1 could recover the proliferation of HSPCs in Cu-stressed embryos (A) Ectopic expression of tuba1a and stmn4 mRNA could recover the expression of runx1 or cmyb in Cu-stressed embryos at 33 hpf. A13, A14, calculation of gene expression in each embryo from different groups. (B) Ectopic expression of foxm1 mRNA could recover the expression of runx1 or cmyb in Cu-stressed embryos at 33 hpf. B9, B10, calculation of gene expression in each embryo from different groups. (C) Ectopic expression of foxm1 could recover BrdU cell proliferation in Cu-stressed embryos at 33 hpf (C1-C12) and 72 hpf (C13-C24). C25, C26, calculation of HSPC proliferation in embryos from different groups. (D) Ectopic expression of foxm1 mRNA could recover the expression of tuba1a and stmn4 in Cu-stressed embryos at 33 hpf. D9, D10, calculation of tuba1a and stmn4 expression in embryos from different groups. (E) Cu significantly suppressed the transcriptional activities of both human (E1) and zebrafish (E2) TUBA1A and STMN4 promoters. (F) Ectopic expression of human FOXM1 significantly up-regulated the transcriptional activities of TUBA1A and STMN4 promoter and Cu overload significantly suppressed the up-regulated transcriptional activities. A1-A12, B1-B8, C1-C24, D1-D8, lateral view, anterior to the left, and dorsal to the up. Data are mean ± SD. t-test, ?p < 0.05, ??p < 0.01, ???p < 0.001. Scale bars, 20 ?m (C1-C24) and 50 ?m (A1-A12, B1-B8, D1-D8). |
|
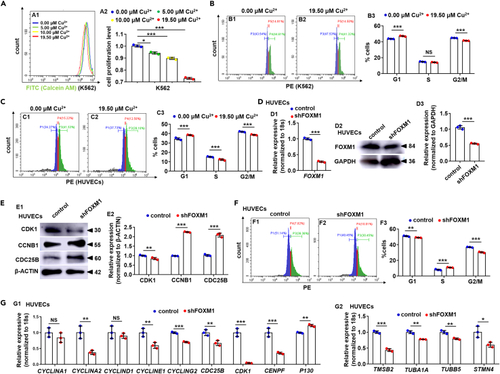
Cu overload impairs the cell cycle in mammalian cells (A) Analysis of cell proliferation in the 0.00 ?M Cu, 5.00 ?M Cu, 10.00 ?M Cu, and 19.50 ?M Cu-stressed K562 cells. A1, flow cytometry (FACS) histogram; A2, calculation of cell proliferation in different groups. (B) Cell cycle in 19.50 ?M Cu-stressed K562 cells. B1, B2, flow cytometry (FACS) histogram; B3, calculation of cell cycle stage. (C) Cell cycle in 19.50 ?M Cu-stressed HUVECs cells. C1, C2, flow cytometry (FACS) histogram; C3, calculation of cell cycle stage. (D) FOXM1 mRNA (D1) and the protein level (D2-D3) were decreased in FOXM1 knockdown (shFOXM1) HUVECs, GAPDH was used as an internal control. (E) Protein levels of cell cycle regulators CDK1, CCNB1, and CDC25B in FOXM1 knockdown (shFOXM1) HUVECs (E1), GAPDH was used as an internal control. (F) shFOXM1 HUVEC cells were blocked at the G1 stage. F1, F2, flow cytometry (FACS) histogram; F3, cell cycle stage calculation. (G) Expression of cell cycle genes (G1) and cytoskeleton genes (G2) in shFOXM1 cells. Data are mean ± SD. t-test, ?p < 0.05, ??p < 0.01, ???p < 0.001. NS, not significant. |
|
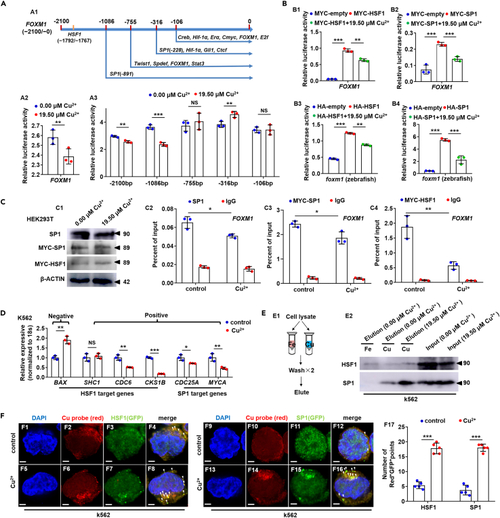
Cu overload impairs HSF1/SP1 transcriptional activities on FOXM1 (A) Schematic diagram of transcription factor targeted domains in FOXM1 promoter (A1), Cu overload significantly suppressed the transcriptional activities of FOXM1 promoters (A2) and transcriptional activities of the 5? truncated promoters of gene FOXM1 (?2100/+28, ?1086/+28, ?755/+28, ?316/+28, ?106/+28) (A3). (B) Ectopic expression of both zebrafish and human HSF1 or SP1 up-regulated the transcriptional activities of FOXM1 promoter, and Cu overload inhibited the increased transcriptional activities on human (B1, B2) and zebrafish (B3, B4) FOXM1 promoters, respectively. (C) Protein levels of SP1, MYC-SP1 (SP1), and MYC-HSF1 (HSF1) in Cu-stressed mammalian cells (C1). Reduced binding enrichment of protein SP1 (C2, C3) and HSF1 (C4) on FOXM1 promoter under Cu stresses as revealed by chromatin immunoprecipitation assays (ChIP). Anti-SP1 and Anti-MYC were used for ChIP assays in the control and the Cu-stressed cells, with anti-IgG used as the negative control. (D) Expression of HSF1 targeting genes BAX, SHC1, CDC6, CKS1B, and SP1 targeting genes CDC25A, MYCA in K562 cells. (E) The binding of the indicated proteins (HSF1/SP1) to Cu2+ and Fe3+ was assessed by western blot analysis of eluted proteins from the indicated metal-loaded resins. (F) Cu probe (Red+) and HSF1/SP1 protein (GFP+) double-positive foci were significantly increased in the cytoplasm of Cu overload K562 cells. F17, calculation of the number of Red+GFP+ foci in K562 cells from different groups. Data are mean ± SD. t-test, ?p < 0.05, ??p < 0.01, ???p < 0.001. NS, not significant. Scale bars, 2 ?m (F1-F16). |
|
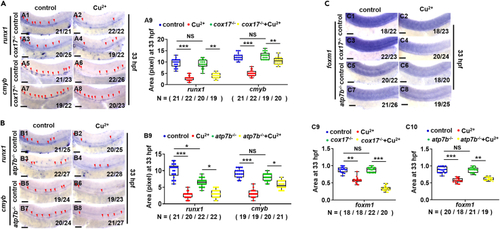
HSPC proliferation in genetic models with dysfunctional Cu homeostasis (A) Expression of runx1 and cmyb in cox17 mutants (cox17?/?) with or without Cu stresses at 33 hpf. A9, calculation of runx1 and cmyb expression in embryos from different groups. (B) Expression of runx1 and cmyb in atp7b mutants (atp7b?/?) with or without Cu stresses at 33 hpf. B9, calculation of runx1 and cmyb expression in embryos from different groups. (C) Expression of foxm1 in cox17 mutants and atp7b mutants with or without Cu stresses at 33 hpf. C9, C10, calculation of foxm1 expression in embryos from different groups. A1-A8, B1-B8, C1-C8, lateral view, anterior to the left, and dorsal to the up. Data are mean ± SD. t-test, ?p < 0.05, ??p < 0.01, ???p < 0.001, NS, not significant. Scale bars, 50 ?m (A1-A8, B1-B8, C1-C8). |