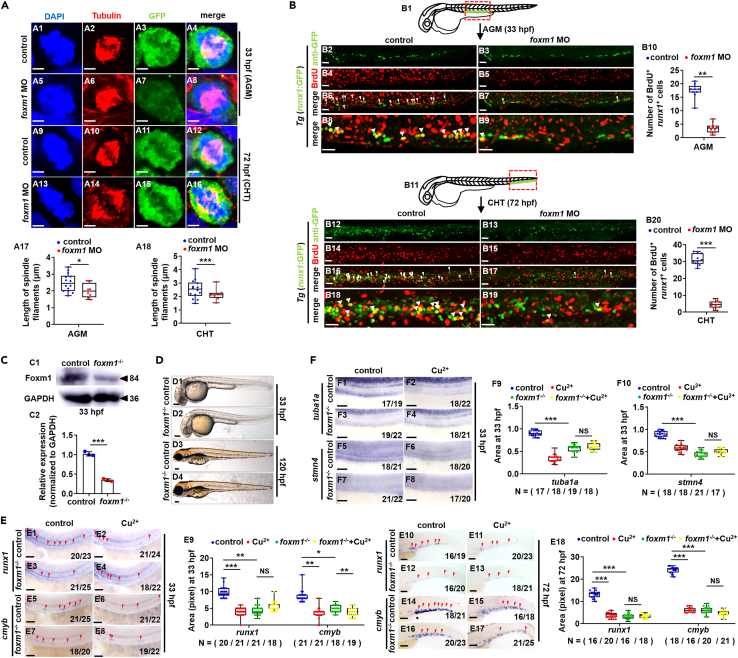
Figure 5
Dysfunction of gene foxm1 impairs the proliferation of HSPCs
(A) Mitotic malformation of HSPCs in foxm1 morphants at 33 hpf (AGM) and 72 hpf (CHT), respectively. A1, A5, A9, A13, DAPI staining; A2, A6, A10, A14, anti-α-tubulin staining; A3, A7, A11, A15, anti-GFP staining; A4, A8, A12, A16, merged. At least 10 mitotic HSPCs in more than 10 embryos were observed for each group. A17, A18, calculation the length of spindles in metaphase runx1GFP+ cells at 33 hpf and 72 hpf, respectively.
(B) BrdU cell proliferation in embryos at 33 hpf (B2-B9) and 72 hpf (B12-B19), respectively. B1, B11, AGM and CHT domain in embryos, respectively; B10, B20, calculation of HSPC proliferation in embryos from different groups.
(C) The protein level of Foxm1 in foxm1 mutants with a 10-bp deletion at 33 hpf. GAPDH was used as an internal control.
(D) Phenotypes of foxm1 mutants at 33 hpf and 120 hpf, respectively.
(E) Expression of runx1 and cmyb in tuba1a mutants with or without Cu stresses at 33 hpf and 72 hpf. E9, E18, calculation of runx1 and cmyb expression in embryos from different groups.
(F) Expression of tuba1a and stmn4 in foxm1 mutants with or without Cu stresses at 33 hpf. F9, F10, calculation of tuba1a and stmn4 expression in embryos from different groups. B2-B9, B12-B19, D1-D4, E1-E8, E10-E17, F1-F8, lateral view, anterior to the left, and dorsal to the up. Data are mean ± SD. t-test, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, NS, not significant. Scale bars, 2 μm (A1-A16), 20 μm (B2-B9, B12-B19), 50 μm (E1-E8, F1-F8), 100 μm (D1-D4) and 200 μm (E10-E17).