- Title
-
Non-nuclear Pool of Splicing Factor SFPQ Regulates Axonal Transcripts Required for Normal Motor Development
- Authors
- Thomas-Jinu, S., Gordon, P.M., Fielding, T., Taylor, R., Smith, B.N., Snowden, V., Blanc, E., Vance, C., Topp, S., Wong, C.H., Bielen, H., Williams, K.L., McCann, E.P., Nicholson, G.A., Pan-Vazquez, A., Fox, A.H., Bond, C.S., Talbot, W.S., Blair, I.P., Shaw, C.E., Houart, C.
- Source
- Full text @ Neuron
|
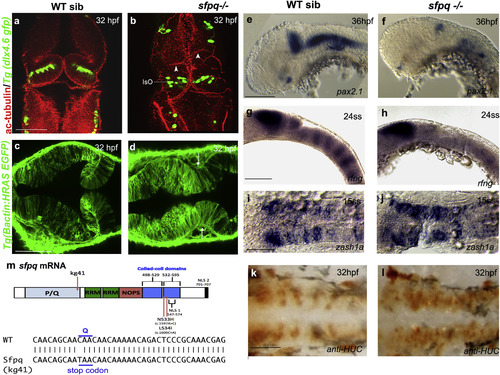
SFPQ Is Required for Brain Boundaries Dorsal (A?D and I?L) and lateral (E?H) views of 32 hpf zebrafish brain with anterior to the top (A and B) or left (C?L). (A and B) Immunostaining of sfpq; Tg(dlx4.6 GFP) embryos. Anti-acetylated tubulin staining (red) reveals asymmetrical folds in the midbrain (white arrowheads) and thickening of the isthmic organizer (IsO) in coma mutant (B; n = 8) compared to its wild-type sibling (A; n = 24). GFP staining (green) reveals disorganized neuronal distribution in the cerebellum. (C and D) Dorsal views, anterior to the top, of sibling (C) and mutant (D) sfpq;Tg(?actin: HRAS gfp) embryos showing failure of morphological thinning of the isthmus (white arrows in D, n = 9/9) in the homozygous mutant. (E and F) Expression of pax2.1 in the MHB, greatly reduced in all sfpq mutants (F; n = 10) compared to siblings (E; n = 30) at 36 hpf. (G and H) Expression of boundary marker, rfng, in siblings (G; n = 21) and mutant (H; n = 7, reduced or absent) at 24 ss. (I and J) Expression of zash1a in the hindbrain at 15 ss, in siblings (I; n = 19) and mutant (J; n = 6). (K and L) HuC staining at 32 hpf, in siblings (K; n = 17) and mutant (L; n = 5). Scale bar, 100 ?m. (M) Schematic of the sfpq gene and the zebrafish and human mutations described in this report and, below, the sequence altered in the zebrafish coma mutant. <>PQ, proline (P) glutamine (Q) rich; RRM, RNA recognition motif; NOPS, NONA/paraspeckle domain; NLS, nuclear localization signals.EXPRESSION / LABELING:
PHENOTYPE:
|
|
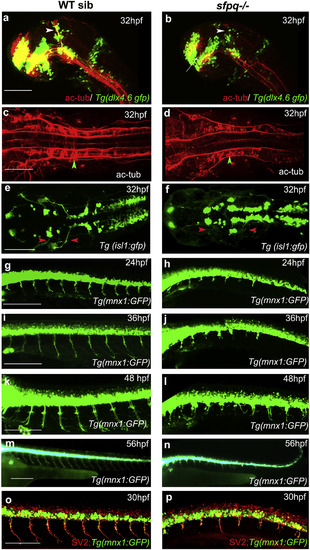
Axonogenesis Is Affected in sfpq?/? Embryos Lateral (A, B, and G?P) and dorsal (C?F) views, anterior to the left, of zebrafish brain at 32 hpf (A and B) and spinal cord at 24?56 hpf. (A and B) sfpq;Tg(dlx4.6:gfp) embryos. Lack of supra-optic commissure (white arrow) and posterior commissure (white arrowhead) in coma (n = 8/32) is revealed by acetylated tubulin staining. (C and D) Dorsal view, anterior to the left of acetylated tubulin staining showing hindbrain disorganized axonal tracks in sfpq mutant (D; green arrowhead, n = 8) compared to siblings (C; n = 26). (E and F) Dorsal view, anterior to the left of GFP+ motor neurons in sfpq; Tg(isl1:gfp) siblings (E) and mutant (F), showing cranial motor neuronal clusters lacking axonal projections in the mutant (red arrowhead, n = 7). (G?N) Lateral view, anterior to the left, of confocal live imaging, showing temporal defect in axonogenesis in the majority of spinal motor neurons in a mutant (H, J, L, and N; n = 14) compared to a sibling (G, I, K, and M; n = 42) in the Tg(mnx1: gfp) background. (O and P) Lateral view, anterior to the left, of SV2 antibody staining of siblings (O) and sfpq null mutant (P), showing pre-synaptic protein in the few axons formed in sfpq mutants (n = 9). Scale bar, 100 ?m. |
|
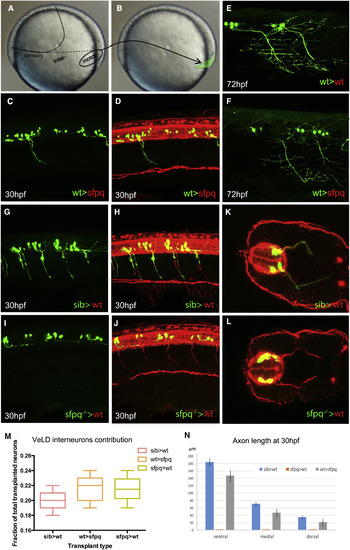
Sfpq Is Required Cell Autonomously for Motor Axon Development Homotopic transplantation at 70%?80% epiboly of sfpq?/?; Tg(mnx1:GFP) and sibling Tg(mnx1:GFP) ventral spinal cord progenitors into wild-type or sfpq?/? hosts. (A and B) Schematic of the transplants from donor (A) to host (B) embryo. (C, D, and G?L) Lateral view, anterior to the left (C, D, and G?J) or transverse (K and L) of 30 hpf zebrafish transplanted trunks, showing the transplanted mnx1+ neurons in green and all axons in red (acetylated tubulin staining). (E and F) Lateral view, anterior left of transplanted wild-type MNs (green) in early larvae (3 dpf) in wild-type (E) or sfpq?/? (F) hosts. (M) Quantification of VeLD interneurons in transplanted clones at 30 hpf (value is number of interneurons/total number of transplanted neurons, number of embryos in box). (N) Quantification of axonal lengths of transplanted neurons at 30 hpf. Scale bar, 100 ?m. |
|
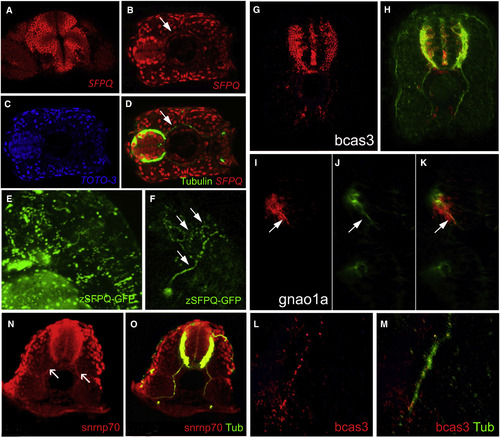
SFPQ Protein Is Found in Axons, with Unprocessed RNA Targets and Spliceosome Component U1 snRNP70 Transverse single confocal sections of 24 hpf (A, N, and O) and 48 hpf (B?M) forebrain (A and I?K) or spinal cord dorsal to the top (G, H, L, and M) or to the left (B?D), and lateral view of head, anterior to the left (E and F). (A?F) SFPQ protein expression shown by antibody staining (A?D) or GFP-tagged ectopically expressed protein (E and F). Identity of red, green, and blue staining is labeled on bottom right corner of each picture. Arrow shows axonal localization. (G?K) Localization of first alternative introns (in red) of SFPQ-dependent transcripts, bcas3 (G and H), and gnao1a (I?K). Axons are stained by acetylated tubulin antibody in green. (L and M) High-resolution imaging of bcas3 intron detection (L; red) in motor axons stained with acetylated tubulin antibody (M; green). (N and O) Double antibody staining of U1 snRNP70 proteins (N) with acetylated tubulin (O; merged channels) in spinal axons. Arrows are pointing to motor axons. Scale bar, 25 ?m. |
|
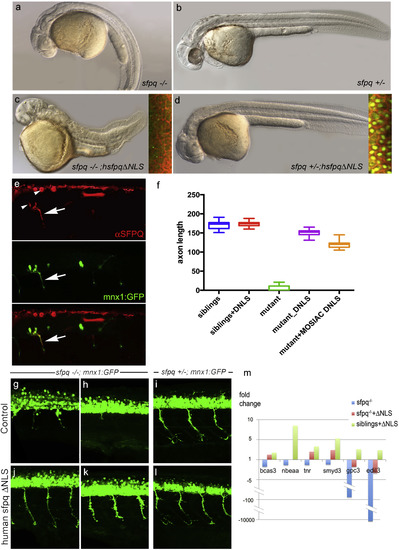
Non-nuclear Sfpq Rescues Loss of Transcripts and Axonal Phenotype in the coma Mutant Lateral view of 30 hpf zebrafish (A?D) and spinal cord (G?L), anterior to the left. (A?D) General morphology of sfpq?/? (A); sfpq+/? siblings (B); sfpq?/? injected with GFP-hsfpq?NLS (C) and sfpq+/? injected with GFP-hsfpq?NLS (D). Insets showing localization of the human GFP-tagged protein in early ss embryos, CAAX-Cherry in red. (E) Whole-mount detection of SFPQ (red) in a 28 hpf sfpq?/? mutant, injected at one-cell stage with pmnx1:GFP and pCS2:hsfpq?NLS DNA, expressing these mosaically. Arrow shows motor axon. Note the negative nuclei (arrowhead). (F) Measurement of ventral motor axon length in siblings and homozygous mutant uninjected or injected with RNA (DNLS) or DNA (MOSAIC DNLS) coding for hSFPQ?NLS. For the controls and RNA injections, the ventral motor axons were measured in five embryos over five-somite length upstream of the cloaca. For mosaic DNA-injected sample, measures were done only in seven homozygous mutants, in the same trunk area for the rare SFPQ+ neurons. (G?L) Lateral view, anterior to the left of spinal cords, showing rescue of the axonal defect both in the anterior (G and J) and posterior (H and K) trunk of sfpq?/?;Tg (mnx1:GFP) by injection of the ?NLS human SFPQ. Sibling axons are unaffected by the injection (I and L). (M) qPCR results for six out of ten DES transcripts from 32 hpf siblings and sfpq?/? mutant embryos uninjected or injected with human ?NLS sfpq RNA. Bars show expression fold changes compared to the expression level of the transcript in uninjected sibling embryos taken as reference. The four transcripts not plotted did not show any improvement after ?NLS rescue. |
|
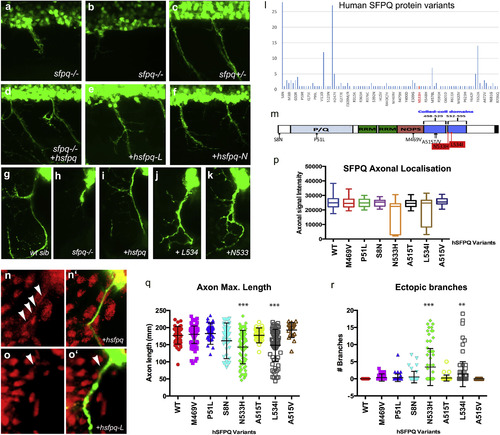
Human Mutations Affect SFPQ Localization and Motor Development in the Zebrafish (A?K) Lateral (A?F) and transverse (G?K, N, and O) views of the spinal cord in sfpq?/? mutant (A, B, and H), siblings (C and G), and homozygous mutant rescued by wild-type human Sfpq (D and I) or L534 (E, J, and O) or N533 (F and K) mutant human Sfpq. (L) Distribution of SFPQ variants in all exome-sequenced human individuals. In red, the position of the variants only found in fALS patients. (M) Schematic of the wild-type hSFPQ protein and the seven variants cloned and tested in double-blind rescue experiments in the Tg(mnx1:GFP) background. S8N (c.23G>A) is present in 4 SALS and 24 ExAC samples, showing a modest enrichment in SALS (p = 0.029, two-tailed Fisher?s test). P51L (c.152C>T) is private to a single SALS patient and absent from all controls. M469V (c.1405A>G) is in a single SALS patient and twice in ExAC (p = 0.071). A515V (c.1544C>T) and A515T (c.1543G>A) are in two and one ExAC normal individuals, respectively, and are located in the coiled-coil domain close to the two FALS variants we identified. (N and O) Close-up of the motor axon and its environment, stained for SFPQ in red and GFP in green in the sfpq?/?; Tg(mnx1:GFP) rescued by injection of the human (N and N?) or L-mutated form (O and O?). Arrowheads show localization of SFPQ in motor axons. (P) Quantification of the ?-SFPQ red fluorescent signal in motor axons on confocal stacks, for five pairs of motor axons per 48 hpf embryo stained with SFPQ antibody. Measurement was done in no less than 30 (maximum, 46) embryos per variant injected. Injection is done in sfpq+/?;Tg(mnx1:GFP) incross progeny. The embryos showing poor signal intensity were genotyped and were all homozygous mutants. (Q) Quantification of the length of ventral motor axons. Measures were made for five segments per 48 hpf embryo (five somites anterior to the cloaca) on confocal stacks, using FIJI Single Neurite Tracer in no less than 28 (maximum, 41) embryos per variant injected. Injection is done in sfpq+/?;Tg(mnx1:GFP) incross progeny. The embryos showing shorter axon length were genotyped and were all homozygous mutants. Asterisks indicate highly significant reduction in axon lengths compared to wild-type (pairwise ANOVA, ???p < 0.0001). The two significantly different variants show same difference when comparing pairwise to the other variants. All other variants are not significantly different from wild-type. (R) Quantification of ectopic ventral motor branching. Measures were made for five segments per 48 hpf embryo (five somites anterior to the cloaca) on confocal stacks using FIJI Single Neurite tracer in no less than 28 (maximum, 41) embryos per variant injected. Injection is done in sfpq+/?;Tg(mnx1:GFP) incross progeny. The embryos showing excessive branching were genotyped and were all homozygous mutants. Asterisks indicate significance (pairwise ANOVA, ???p < 0.0001 and ??p < 0.001) compared to wild-type. The two significantly different variants show same difference when comparing pairwise to the other variants. All other variants are not significantly different from wild-type. |

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. PHENOTYPE:
|

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. PHENOTYPE:
|
|
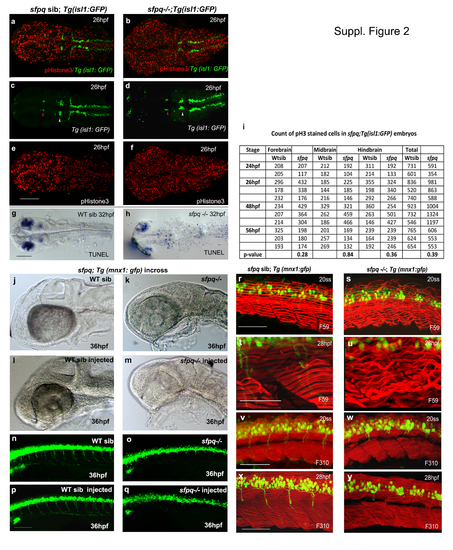
(Related to Figure 1) a. Temporal phenotypic progression in sfpq mutant Lateral view of live WT siblings and sfpq mutant with anterior to the left at various developmental stages: 28hpf (n=31), 30hpf (n=47), 38hpf (n=21), 48hpf (n=19), 52hpf (n=41) and 58hpf (n=71), with 25% of the total (n) are homozygous mutants . Scale bar=100?m. b. Normal overall brain patterning and induction of MHB in sfpq mutant Lateral view of zebrafish brain (a-l) and spinal cord (m,n,q,r) with anterior to the left. Normal induction of MHB (black arrowhead) is observed in sfpq mutants with normal expression of pax2.1 and fgf8 at the MHB at 15ss (n=55; n=62). Expression of fgf8 and foxb1.2 in the MHB (white arrowhead) is greatly reduced in the sfpq mutants ( n=10/40; n=5/21) by end of somitogenesis. No difference in the pattern of netrin1 and lhx5 expression was detected (n=9/36; n=6/25). c. Wnt signaling in the sfpq mutant The axin2 and topd gfp expression was normal in the spinal cord and brain except for its expression (white arrow) in the hypothalamus of WT siblings that is absent in the sfpq-/- embryos (n=11/42; n=7/35) showing the absence of Wnt activity in the region. Scale bar=100?m. c. Rescue of mutant phentotpye with sfpq RNA injection Dorsal view (a-d) of zebrafish brain at 30hpf and lateral view (e-h) of zebrafish spinal cord at 32hpf with anterior to the left. The embryos from both the sfpq;Tg(isl1:gfp) incross (n=51) and the sfpq;Tg(mnx1:gfp) incross (n=76) injected with sfpq mRNA appeared almost normal compared to its control WT siblings with the rescue of axonogenesis defect and the kinky tail that is observed in the non-injected sfpq mutants. Scale bar=100?m. |
|
(Related to Figure 2). Cell proliferation and apoptosis in sfpq mutant Dorsal view of zebrafish brain at 26hpf (a-f) and 30hpf (g-h) with anterior to the left. a-f. Cell proliferation in sfpq mutant brain (n=11) analysed using gfp expression in sfpq;Tg:isl1 gfp as a landmark to delineate the domains: nIII neurons (red arrowhead ) as posterior limit of the forebrain and the anterior limit of the midbrain; nV neurons (white arrowhead ) as the posterior limit of the midbrain and the anterior limit of the hindbrain. Cell proliferation in the tail of sfpq mutant was found in par with that of its WT siblings (data not shown). g,h. Increased apoptotis is observed throughout the the sfpq brain (n=4) compared to siblings (n=12). i. Proliferation quantified using phosphor-Histone3 staining, counted in different brain regions of sfpq mutant and siblings. No significant difference was observed in cell count (Student?s t-test, p>0.05). Increased cell proliferation observed in some (n=5 of the 11 counted) may be due to cells getting arrested at G2/M phase of cell cycle. sfpq initial phenotype is independent of cell death j-q. p53 knockdown in sfpq;Tg(mnx1: gfp) embryos. Lateral view of zebrafish brain (jm) and spinal cord (n-q) at 36hpf with anterior to the left. The p53 MO injected sfpq-/- embryos (k, o. n=3) show similar MHB phenotype and axonogenesis defect as the noninjected mutants (m, q. n=5), compared to injected (l, p. n=14) and non-injected siblings (j, n. n=11). Scale bar=100?m. Muscle formation in sfpq mutant r-y. Lateral view of zebrafish muscle, anterior to the left. r-u. Immunostaining for slow muscle with F59 antibody shows proper somatic boundaries and differentiated slow muscle fibres and sarcomeres in the sfpq mutant (20ss: n=6/25; 28hpf: n=9/39). The wavy appearance of slow muscle fibres in the mutant at later stages compared to its WT siblings (c-f), likely due to inactivity. v-y. Immunostaining for fast muscle with F310 antibody shows the integrity of the fast muscle in sfpq mutant (20ss: n=5/22; 28hpf: n=6/24) as normal as in its WT siblings. Scale bar=100?m. |

Unillustrated author statements PHENOTYPE:
|