- Title
-
Mutations in zebrafish pitx2 model congenital malformations in Axenfeld-Rieger syndrome but do not disrupt left-right placement of visceral organs
- Authors
- Ji, Y., Buel, S.M., Amack, J.D.
- Source
- Full text @ Dev. Biol.
|
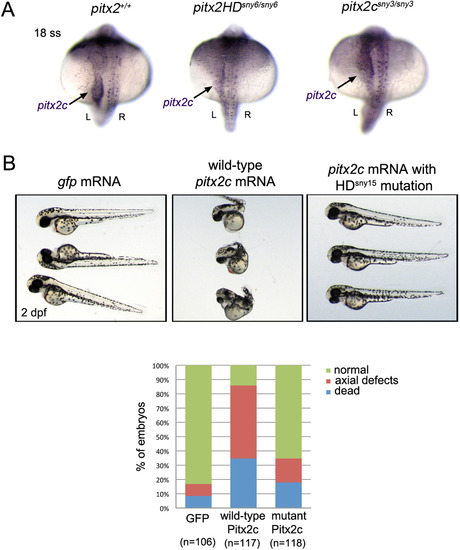
Functional analysis of mutant pitx2HD transcripts. (A) RNA in situ hybridizations revealed normal pitx2 mRNA expression in wild-type, pitx2HD-/- and pitx2c-/- embryos, including asymmetric pitx2c in lateral plate mesoderm (arrow) at the 18 somite stage (18 ss). L=left; R=right. (B) Microinjection of wild-type pitx2c mRNA into wild-type embryos resulted in severe developmental defects or lethality at 2 dpf, which was not frequently observed in controls injected with gfp mRNA. Most embryos injected with pitx2c mRNA containing the HDsny15 mutation developed normally. n=number of embryos analyzed (pooled from three experiments). EXPRESSION / LABELING:
|
|
Pitx2HD mutants grow to adulthood and develop eye defects. (A) Normal gross appearance of wild-type, pitx2HDsny7/sny7 and pitx2csny3/sny3 fish at 5 dpf. (B) Homozygous pitx2HD mutants reached adulthood, but were smaller than heterozygous siblings and had malformed eyes (arrow). Adult homozygous pitx2c mutants were indistinguishable from wild-type siblings. (C) Eye and craniofacial phenotypes in adult fish. Ocular defects of the iris and cornea (arrow) were observed in pitx2HD mutants, but not pitx2c mutants. |
|
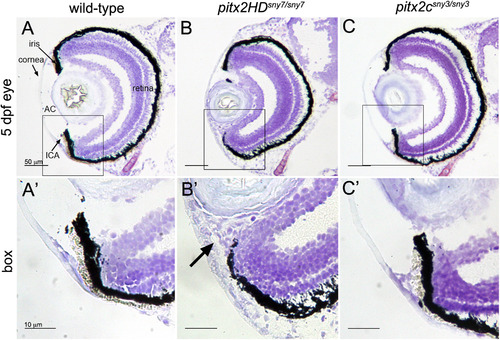
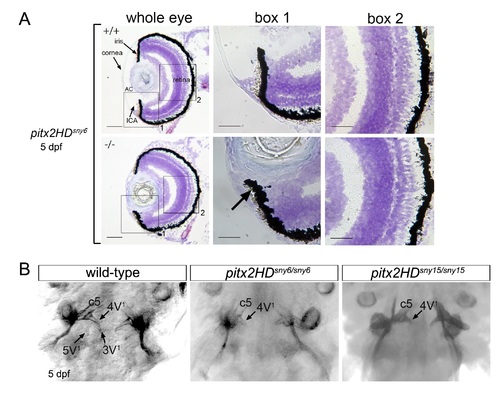
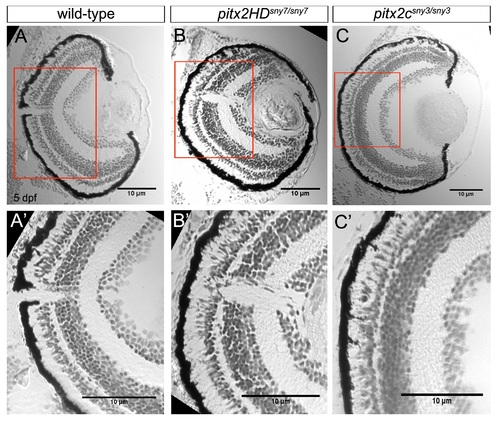
Malformations of the anterior segment of the eye in Pitx2HD mutant embryos. (A-C) Cryosections of the eye from 5 dpf wild-type (A), pitx2HDsny7/sny7 (B) and pitx2csny3/sny3 (C) fish stained with crystal violet. AC=anterior chamber; ICA=iridocorneal angle. Scale bars=50 Ám. (A′-C′) Enlarged view of box 1 in A-C showing anterior segment structures. In pitx2HD mutants (B′), the anterior chamber was reduced and the iridocorneal angle (arrow) was malformed. Scale bars=10 Ám. PHENOTYPE:
|
|
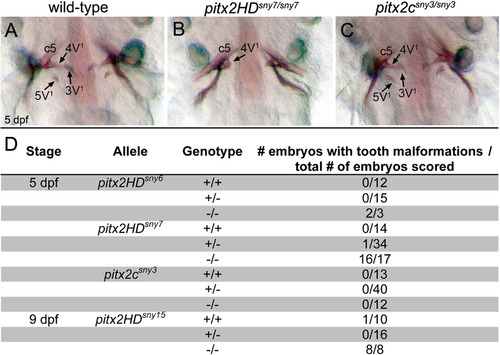
Tooth morphogenesis is disrupted in Pitx2HD mutants. (A-C) Ventral views of alizarin red staining of teeth at 5 dpf in wild-type (A), pitx2HDsny7/sny7 (B) and pitx2csny3/sny3 (C) fish. c5: fifth ceratobranchial; 4V1, 3V1 and 5V1 designate individual teeth. (D) Quantification of tooth defects observed in pitx2HDsny7 and pitx2csny fish at 5 dpf. Tooth malformations were defined as a reduced number of teeth and incomplete tooth formation as shown in (B). PHENOTYPE:
|
|
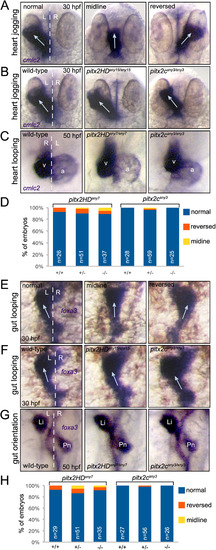
Direction of asymmetric heart and gut morphogenesis is normal in pitx2 mutants. (A-C) RNA in situ hybridizations using cmlc2 probes labels the developing heart tube. (A) Selected wild-type embryos that show the potential outcomes of heart jogging at 30 hpf, which include normal left-sided jogging, jogging along the midline and reversed jogging. (B) Representative images of heart jogging in wild-type and pitx2HD and pitx2c mutant embryos. Arrows indicate direction of heart jogging. (C) Normal asymmetric heart looping in wild-type and pitx2HD and pitx2c mutant embryos placed the ventricle (v) to the right of the atrium (a) at 50 hpf. (D) Heart looping direction was scored as either normal, reversed or along the midline for individual embryos at 50 hpf and the genotype (+/+, +/- or -/-) was then determined for each embryo. (E-G) foxa3 in situ hybridizations marked the developing gut tube. (E) Potential outcomes of gut looping at 30 hpf include normal left-sided looping, remaining at the midline and reversed looping. (F) Direction of gut looping was similar in wild-type and pitx2HD and pitx2c mutant embryos. Arrows indicate direction of gut looping. (G) Normal asymmetric placement of the liver (Li) and pancreas (Pn) at 50 hpf. (H) Gut asymmetry was scored at 50 hpf and then each embryo was genotyped. No statistical differences in heart looping (D) or gut asymmetry (H) were identified using single factor one way ANOVA. White dashed lines represent the midline; L=left; R=right; n=number of embryos analyzed. PHENOTYPE:
|
|
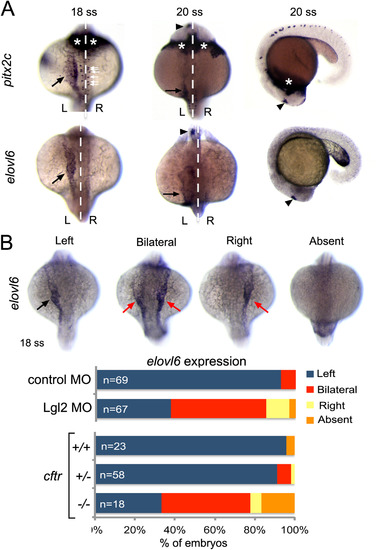
Asymmetric expression of elovl6 depends on Kupffer′s vesicle. (A) RNA in situ hybridizations revealed similar asymmetric expression patterns for pitx2c and evolv6 in left lateral plate mesoderm (black arrows) and brain (arrowheads) at 18-20 somite stages (ss). pitx2c mRNA was detected in hatching gland (asterisks) and neurons (white arrows), whereas elovl6 was not expressed in these domains. White dashed line is the midline; L=left and R=right. (B) Representative images are shown of left-sided, bilateral, right-sided and absent elovl6 expression in lateral plate mesoderm (arrows) at 18 somite stage. The graph shows quantification of asymmetric elovl6 expression profiles in control, Lgl2 depleted and genotyped cftrpd1048 mutant embryos. EXPRESSION / LABELING:
PHENOTYPE:
|
|
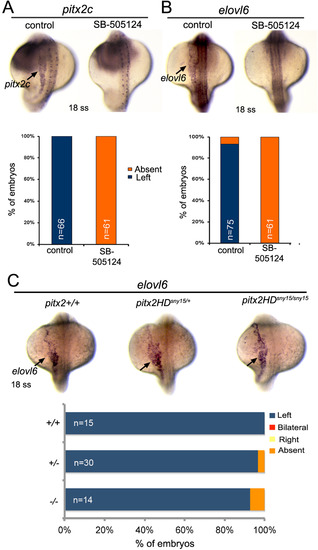
Asymmetric expression of elovl6 requires Nodal signaling but does not depend on Pitx2. (A,B) Analysis of asymmetric pitx2c (A) or elovl6 (B) expression (arrows) at the 18 somite stage (ss) in control embryos and embryos treated with SB-505124 to block Nodal signaling. The graphs indicate the percentage of embryos with normal left-sided expression or absent expression. (C) Asymmetric elovl6 expression (arrows) in wild-type and pitx2HD mutant embryos. n=number of embryos analyzed. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Gross morphology of pitx2HD mutants. (A) Gross appearance of wild-type, pitx2HDsny6/sny6 and pitx2HDsny15/sny15 fish at 5 dpf. (B) Eye phenotypes in adult fish were similar in each of the three HD alleles generated. |
|
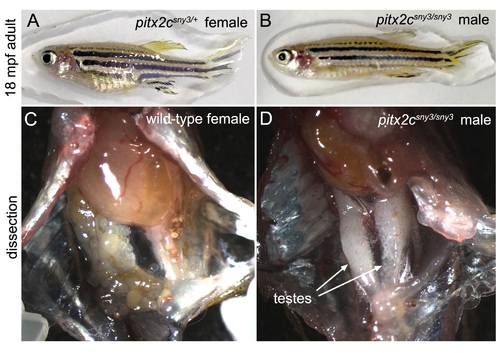
All pitx2csny3/sny3 adults recovered were male. (A-B) Gross appearance of a representative adult heterozygous pitx2c+/sny3 female (A) and a homozygous pitx2csny3/sny3 male (B) at 18 months post-fertilization (mpf). (C,D) Dissections revealed testes in pitx2csny3/sny3 males (n=5/5). PHENOTYPE:
|
|
Pitx2HD mutants develop eye and tooth defects associated with ARS. (A) Cryosections of the eye from 5 dpf wild-type (+/+) or pitx2HDsny6/sny6 (-/-) fish stained with crystal violet. AC=anterior chamber; ICA=iridocorneal angle. Scale bars=50 Ám. Box1 and Box2 are zoom-in views of boxed structures. Retina organization appeared similar in wild-type and pitx2HD mutant fish. (B) Ventral views of alizarin red staining of teeth at 5 dpf wild-type, pitx2HDsny6/sny6 and pitx2HDsny15/sny15 fish. |
|
Retina layers are intact in pitx2 mutants at 5 dpf. (A-C) Cryosections of the eye from 5 dpf wild-type (A), pitx2HDsny7/sny7 (B) and pitx2csny3/sny3 (C) fish stained with crystal violet. (A′-C′) Enlarged view of boxes in A-C showing layers of the retina. Scale bars=10 Ám. Supplementary material |
|
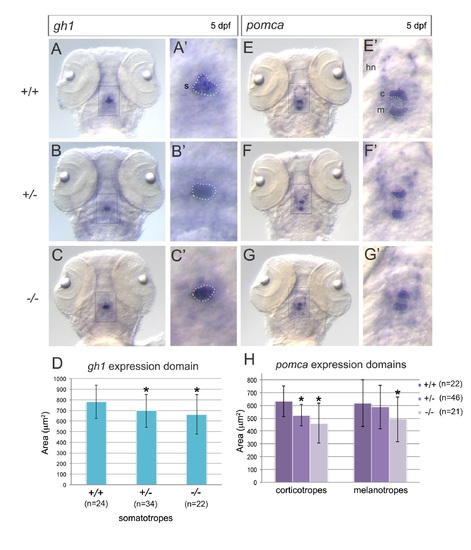
Expression of pituitary markers in pitx2HD mutants. (A-C, E-G) Representative images of RNA in situ hybridizations of gh1 expression (A-C) and pomca expression (E-G) in wild-type (A, E;+/+), heterozygous (B, F;+/-) and homozygous mutant (C, G; -/-) pitx2HDsny15 embryos at 5 dpf. A′-C′ and E′-G′ are enlarged views of boxed areas in A-C and E-G, respectively. s=somatotrope domain; c=corticotrope domain; m=melanotrope domain; hn=hypothalamic neurons. Dashed lines indicate expression domains that were quantified. (D, H) Quantification of the area of the gh1 expression domain (D) and pomca expression domains (H) in pitx2HDsny15 embryos. *=significant difference (p<0.05) from wild-type using Student?s T-test. n=number of embryos analyzed. Supplementary material |
|
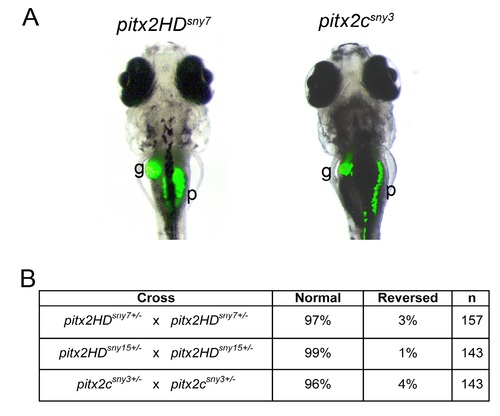
Normal asymmetric placement of visceral organs in pitx2 mutants. (A) Representative images of auto-fluorescence in the gall bladder (g) and pancreas (p) of 7 dpf larvae. L=left; R=right. (B) Scoring of organ placement in larvae from pitx2 heterozygous parents (25% expected to be homozygous mutants). n=number of embryos analyzed. |
|
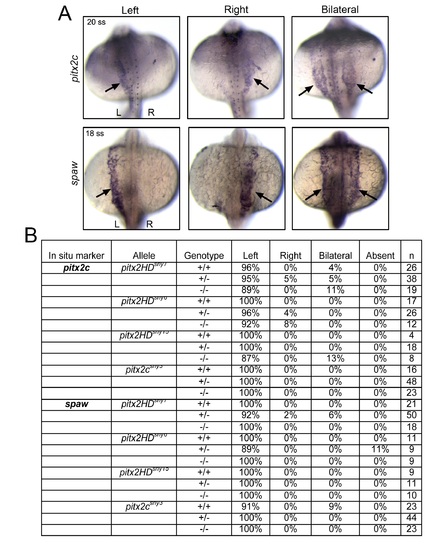
Asymmetric expression of spaw and pitx2c is normal in pitx2 mutants. (A) RNA in situ hybridizations of pitx2c (20 somite stage) and spaw expression (18 somite stage). Representatives of left-sided, right-sided or bilateral expression in lateral plate mesoderm (arrows) are shown. (B) Normal left-sided expression of pitx2c and spaw was observed in most pitx2HD and pitx2c embryos. All embryos were genotyped following in situ hybridization analysis. |
|
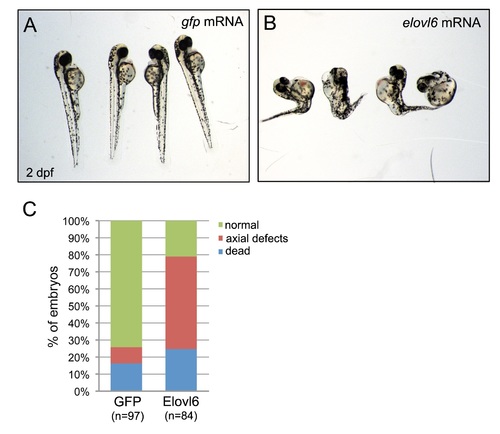
Elovl6 gain-of-function experiments. (A,B) Microinjection of full-length elovl6 mRNA into wild-type embryos resulted in severe developmental defects or lethality at 2 dpf (A), which was not frequently observed in controls injected with gfp mRNA (B). (C) Quantification of defects observed in injected embryos. n=number of embryos analyzed (pooled from three experiments). Supplementary material |

Unillustrated author statements |
Reprinted from Developmental Biology, 416(1), Ji, Y., Buel, S.M., Amack, J.D., Mutations in zebrafish pitx2 model congenital malformations in Axenfeld-Rieger syndrome but do not disrupt left-right placement of visceral organs, 69-81, Copyright (2016) with permission from Elsevier. Full text @ Dev. Biol.