- Title
-
Simplet controls cell proliferation and gene transcription during zebrafish caudal fin regeneration
- Authors
- Kizil, C., Otto, G.W., Geisler, R., Nüsslein-Volhard, C., and Antos, C.L.
- Source
- Full text @ Dev. Biol.
|
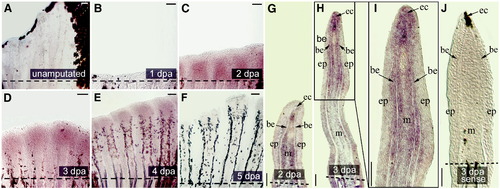
Expression of smp in the regenerating adult caudal fin. smp expression at different time points of caudal fin regeneration are shown from whole mounts (A?F) and coronal cryosections (G?J). (A) The gene is not expressed in unamputated (B) nor in regenerating fins 1 day post amputation (dpa). (C) By 2 dpa, smp is transcribed in the regenerating tissue distal to the amputation plane. Expression localizes to the distal portion of the regenerate by 3 dpa (D) as well as 4 dpa (E). (F) Transcript is down-regulated by 5 dpa. (G) In situ hybridization on cryosections of 2 dpa regenerating fin: smp expression in the mesenchyme (m) underlying the epithelial cap (ec) and basal epithelium (be) of the epidermis. (H) By 3 dpa, smp expression is located several tissues in the distal portion of the regenerate. (I) Higher magnification of distal fin regenerate. (J) Hybridization on 3 dpa fin with sense probe. The dashed line shows the amputation plane. Scale bars equal 100 μm. EXPRESSION / LABELING:
|
|
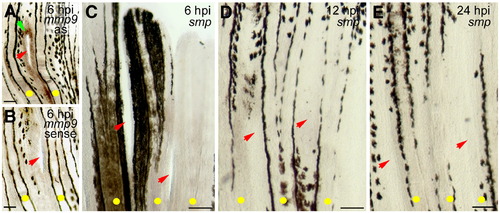
smp transcription is not activated during wound healing. (A) Small 0.2?0.7 mm incisions (red arrows) between the bony fin rays (yellow dots) result in a wound closure response associated with the activation of mmp9 (green arrowhead), a gene activated during wound healing, by 6 h post injury (hpi). (B) Sense control of mmp9 lacks any staining. (C) smp gene transcription is not detected at 6 hpi, (D) 12 hpi, or (E) 24 hpi. By 24 hpi, the superficial wound incision has healed. All scale bars equal 100 μm. EXPRESSION / LABELING:
|
|
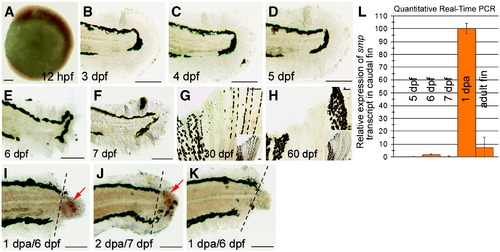
smp expression during zebrafish caudal fin development. Whole-mount in situ for smp in the zebrafish embryo (A), larval fin folds (B?F), juvenile fins (G and H) and regenerating larval fins (I?K). smp expression is in the head and trunk, and reduces towards the posterior at 12 hpf (A). Larval caudal fins show lack of expression at 3 dpf (B), 4 dpf (C), 5 dpf (D), 6 dpf (E) and 7 dpf (F). smp expression is not detected in developing juvenile fin at 30 dpf (G) [insert showing entire fin lobe] and 60 dpf (H) [insert showing entire fin lobe]. (I) Amputation injury at 5 dpf causes up-regulation of smp in the regenerating tissue of the larval caudal fin by 1 dpa (6 dpf). (J) Expression continues at 2 dpa (7 dpf). (K) Sense control of regenerating caudal finfold of the larva. Dashed lines show amputation plane. Scale bars equal 50 μm (A) and 100 μm (B?K). (L) Quantitative RT-PCR for smp message expression in posterior of zebrafish larvae at indicated developmental time points. smp expression is strongly upregulated upon amputation injury at the regenerating larval fin at 1 dpa. EXPRESSION / LABELING:
|
|
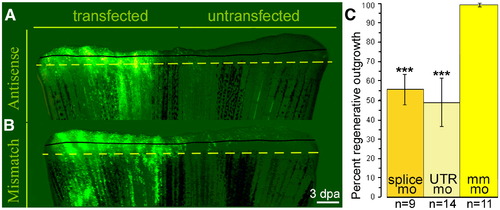
smp is necessary for regenerative outgrowth. (A) Lobes transfected with smp antisense morpholino showed strong fluorescein labeling. (B) Transfection of mismatch morpholino. Solid black lines show extent of regeneration before transfection and dashed lines show level of amputations. Scale bar equals 200 μm. (C) The graph represents the proportional relationship between transfected and untransfected lobes from each fin. n = number of fins transfected and analyzed. ***: p-value < 0.001, Student's t-test. PHENOTYPE:
|
|
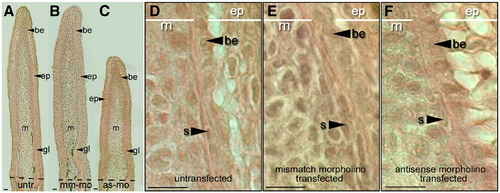
Tissues of the early regenerating fin are still present in morpholino-transfected regenerating fins. (A) Coronal sections through a regenerating caudal fin from a wild-type zebrafish 3 dpa. The regenerative outgrowth of the fin stained with Hematoxylin and Eosin: the epidermis (ep), basal epithelium (be), mesenchyme (m) and growing lepidotrichia (gl). (B) Section through fin lobes transfected with mismatch control morpholinos. (C) Fins transfected with antisense morpholinos. (Dashed lines show amputation plane.) (D) High magnification of coronal section through untransfected regenerating fins show all cell types (s: scleroblasts). (E) High magnification of regenerate transfected with mismatch control morpholino. (F) High magnification of a regenerate transfected with antisense morpholino. Scale bars equal 100 μm (A?C) and 25 μm (D?F). PHENOTYPE:
|
|
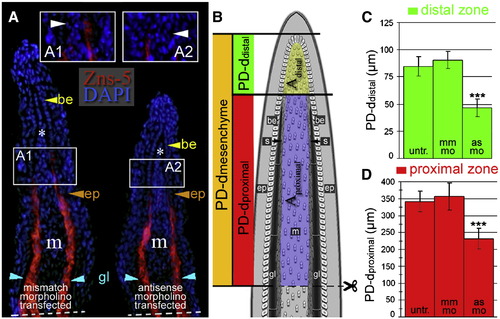
smp knock-down alters morphological proportions in regenerating fin rays. (A) Immunohistochemical labeling of lepidotrichial scleroblasts (red) with the Zns-5 antibody on coronal sections through a regenerating caudal fin from mismatch- and antisense-morpholino-transfected fins 3 dpa. Nuclei were stained with DAPI (blue). Inset panels (A1 and A2) show the most distal end of the Zns-5 labeling: the boundary between distal and proximal zones. Dashed lines show amputation planes. (B) A diagram of a regenerating fin illustrating different regions based on the presence of scleroblasts (s) and Zns-5. (C and D) The graph of the direct measurements of the distal (scleroblast negative) compartment (C) and proximal (scleroblast positive) compartment (D) along the proximodistal axis (PD) between untransfected, mismatch-transfected and antisense-morpholino-transfected fins. Thirty-two cryosections for Zns-5 IHC were measured. (***: p < 0.001, Student's t-test). EXPRESSION / LABELING:
|
|
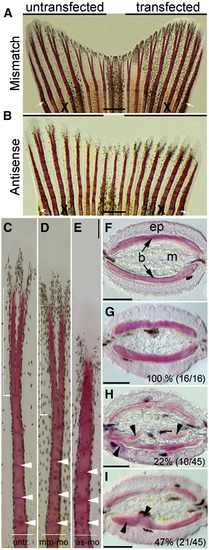
Knockdown of smp results in ectopic bone formation in regenerating fins. (A) Zebrafish caudal fins were allowed to regenerate 5 days after transfecting one lobe with mismatch morpholino (7 dpa). (B) Fin lobes transfected with antisense morpholino. White arrow indicate amputation plane and ?X? indicates the compared rays. (C) Individual rays from untransfected lobes show regular segmentation (white arrow heads) and distal bifurcation of the ray (white square bracket). (D) Rays from mismatch-morpholino-transfected fin. (E) Bony rays from antisense-morpholino-transfected fin lobes with shorter and thicker bone segments (white arrowheads). Distal bifurcation is not visible. (F) Axial cross sections through fin show the epidermis (ep), the two hemi-rays bones (b) and interray mesenchyme (m) from one lepidotrichia. (G) Cross sections through hemi-rays of mismatch-morpholino-transfected fin lobe. Inset number indicates phenotype/total (H) Cross section through hemi-ray of antisense-morpholino-transfected fin lobe shows ectopic bone (black arrow heads). (I) Antisense-morpholino-transfected fins displayed regional thickening within the fin bones (black arrow heads). Scale bars equal 500 μm (A and B) and 100 μm (C?I). PHENOTYPE:
|
|
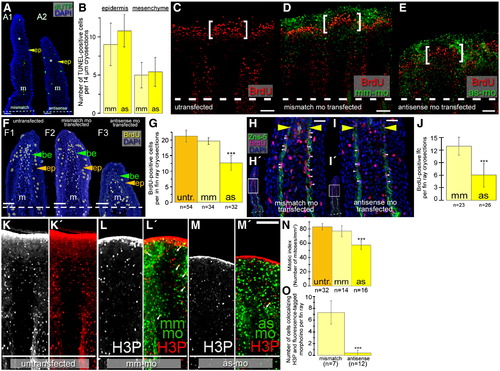
smp knockdown results in reduced BrdU incorporation and histone H3 phosphorylation in cells of the regenerating fin. (A) TUNEL labeling (green fluorescence) of untransfected (A1) and antisense morpholino-transfected (A2) fins. DAPI staining (blue fluorescence) labeled nuclei. (B) Graph shows the average number of TUNEL positive nuclei. Twenty-eight cryosections for TUNEL-assay were analyzed. Scale bar equals 25 μm. Confocal fluorescence imaging of Cy3-labeled BrdU and fluorescein-labeled morpholinos in untransfected (C), mismatch-morpholino-transfected (D), and antisense-morpholino-transfected (E) regenerating fins 3 dpa. BrdU labeling in coronal sections through regenerating lobes of untransfected (F1), mismatch morpholino-transfected (F2) and antisense morpholino-transfected (F3) fins. (G) Graph shows the number of BrdU-positive cells per 14 μm section of untransfected (untr.), mismatch-morpholino-transfected (mm) and antisense-morpholino-transfected (as) fins. (H,I) Immunohistochemistry for the extracellular matrix of scleroblasts (Zns-5, green), Cy3-labeled BrdU-positive nuclei (red) and DAPI-labeled nuclei (blue). Yellow arrowheads mark the most distal localization of Zns-5 staining. White arrowheads highlight BrdU-incorporated scleroblasts. (H′, I′) Panels show entire stained fin and white boxes show region where the larger panels are located. (J) Graph shows the number of Zns-5 and BrdU double positive cells in fins transfected with mismatch and antisense morpholinos. Panels show fluorescent detection of Cy3-labeled histone H3 phosphorylated nuclei alone in untransfected (K), mismatch-morpholino-transfected (L) or antisense-morpholino-transfected (M) or merged with fluorescein-labeled morpholinos: untransfected (L′), mm-mo-transfected (L′), or as-mo-transfected (M′). (N) Graph shows the number of mitoses per mm2 in untransfected (untr.) control, mm-mo-transfected control and as-mo-transfected fins. (O) Graph depicts the number of doubly stained cells for Cy3 (H3P) and fluorescein (morpholino). Scale bars represent 100 μm in C?E and K?M′; 50 μm in F, and 25 μm in H and I. (m: mesenchyme, be: basal epithelium, ep: epithelial layers.) (***: p < 0.001, Student's t-test). |
|
Knock-down of smp results in reduced transcription of cell cycle-associated genes while increased expression of msxb, msxc and shh. (A) The graph of the measurements from real-time quantitative RT-PCR shows fold changes in the transcription of genes associated with the cell cycle between antisense-morpholino-transfected and mismatch-morpholino-transfected regenerating fin lobes. (B) A graph shows fold change expression of several developmental genes after transfection of antisense-morpholino into cells of the regenerating fin. The numbers in parenthesis represent the number of individual replicates. (C) In situ hybridization with msxb probe show expression in untransfected fins. (D) Knock-down of smp expanded the msxb expression domain in the regenerating fin. (E) In regenerating fins of wild-type fish, shh is present in a defined region of basal epithelium that overlies distal margin of new developing scleroblasts. (F?H) shh expression in smp-morphant fins expanded dramatically to the underlying mesenchyme. Three sections from three different experiments are shown. (I) shh expression in untransfected and antisense-morpholino-transfected lobes by whole mount of caudal fins at 3 dpa. (J) The untransfected lobe shows shh expression in the regenerating fin rays as doublets (red arrows). A clearly discernible gap lacking shh transcript is between these doublets. (K) Antisense-morpholino transfected lobe at high-magnification shows a single shh expression domain (red arrows). Dashed lines (black arrows in H) show amputation planes. Scale bars equal 75 μm. (**: p < 0.005; *:p < 0.05 in Student's t-test). |
|
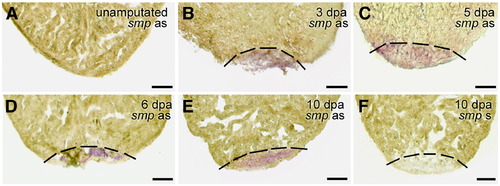
Expression of smp in the regenerating adult heart. (A) Sham operated control hearts lack expression of smp gene. (B) smp gene activity is detectable in the 3 dpa hearts in the ventricular tissue as well as 5 dpa (C), 6 dpa (D) and 10 dpa (E). Sense probe shows no signal (F). Dashed black line indicates the amputation plane. Scale bars equal 100 μm. EXPRESSION / LABELING:
|
|
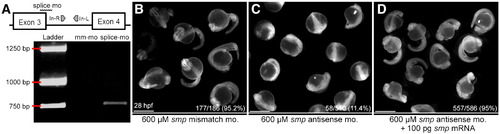
Morpholino-mediated knock-down of smp is specific and can be rescued. (A) Diagram of the genomic region encompassing exons 3 and 4 as well as intron 3 of smp. Symbols mark positions of the antisense splice-junction morpholino (splice mo) and PCR primers (In-R and In-L). Underlying panel is a gel picture with DNA ladder and RT-PCR of mismatch-morpholino-transfected (mm-mo) and antisense-splice-junction-morpholino-transfected (splice mo) embryo extracts. (B) 28 hpf embryos after injection with mm-mo. (C) Embryos at the same stage after injection with splice-mo. (D) Rescued phenotype of same-staged embryos after co-injection of smp mRNA with splice-mo. Numbers indicate embryos showing wild-type phenotype/total injected embryos. The percents are the ratios of observed wild-type phenotype to total. Embryos in (B?D) show emissions from fluorescein-tagged morpholinos. Scale bars equal 500 μm. |
|
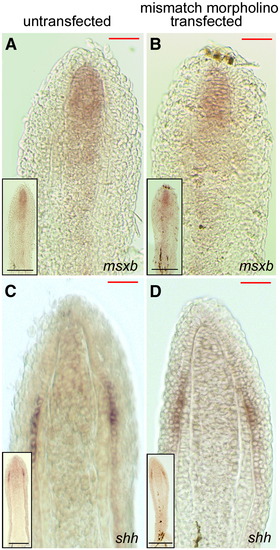
Expression of msxb and shh in regenerating fins is unchanged after transfection with mismatch morpholino. In situ hybridization for msxb (A and B) and shh (C and D) on coronal cyrosections of regenerating fins. The wild-type expression patterns for msxb (A) and shh (C) are unchanged by the transfection of the mismatch morpholino for smp: msxb (B) and shh (D). Insets show the whole section. Scale bars equal 100 μm. |
Reprinted from Developmental Biology, 325(2), Kizil, C., Otto, G.W., Geisler, R., Nüsslein-Volhard, C., and Antos, C.L., Simplet controls cell proliferation and gene transcription during zebrafish caudal fin regeneration, 329-340, Copyright (2009) with permission from Elsevier. Full text @ Dev. Biol.