Figure 8—figure supplement 1—source data 3.
- ID
- ZDB-FIG-240620-251
- Publication
- Hammond et al., 2024 - Tribbles1 is host protective during in vivo mycobacterial infection
- Other Figures
- All Figure Page
- Back to All Figure Page
|
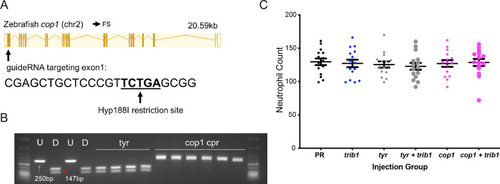
An efficient CRISPR–Cas9 guide RNA was generated for the zebrafish cop1 gene. (A) The zebrafish cop1 gene (ENSDARG00000079329) has one coding transcript which was used as a basis for CRISPR–Cas9 guide design. Exon map and gene information obtained from Ensembl database. The web tool ChopChop was used to design a cop1 guide RNA with suitable restriction enzyme bind site (black line). PAM cut site (NGG) denoted with blue line. (B) Electrophoresis gel of larvae injected with cop1 guide RNA (cpr) with a Hyp188I restriction digest. The undigested PCR band is 250 bp and the successfully digested band is 147 bp. Two uninjected fish were used as controls without (U) and with (D) digestion. Four tyrosinase (tyr) guide injected larvae had conserved SacII sites while 6/6 cop1 larvae did not digest with Hyp188I indicating a genomic disruption at the CRISPR PAM site. First and last lanes of gel images contain NEB Low Molecular Weight ladder for reference. (C) Wholebody Tg(mpx:GFP)i114 neutrophil numbers in 2 dpf larvae injected at the one-cell stage with either phenol red (PR), trib1 RNA overexpression (trib1), tyrosinase CRISPant (tyr), trib1 and tyrosinase CRISPant (tyr + trib1), cop1 CRISPant (cop1), or cop1 and trib1 CRISPant (cop1 + trib1). Data shown are mean ± standard error of the mean (SEM), n = 15 fish per group. |