Fig. S1
|
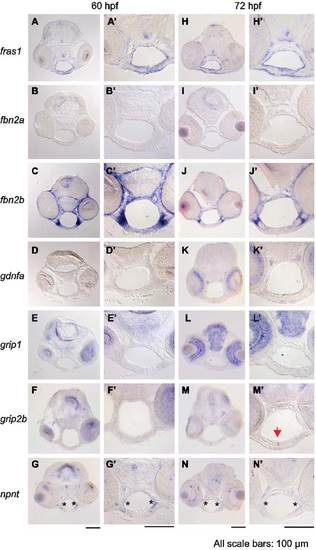
Expression of fras1-related and itga8-related genes in facial tissues. (A-N′) RNA in situ hybridization on wild type tissue sections at (A-G′) 60 hpf or (H-N′) 72 hpf. Each image in A-N shows a lower magnification image of individual sections and A′-N′ shows higher magnification images of these sections, focusing on the pharyngeal region. (A and H) fras1 expression in facial endoderm and ectoderm is shown as a positive control. (B and I) fbn2a shows no detectable facial expression. (C and J) fbn2b is strongly expressed in arch mesenchyme. (D and K) The fras1 processing gene grip1 is expressed in brain and eyes, but not in the pharyngeal regions. (E and L) Similarly, grip1 paralog grip2b is not visibly expressed in pharyngeal regions at 60 hpf (F), but is present by 72 hpf in the joint between the left and right Meckel?s cartilage (M, arrow). This suggests that genes other than grip1 and grip2b may process Fras1 in the face, or that earlier grip expression enables Fras1 processing. Two primer-sets failed to amplify grip2a cDNA at 3 dpf, so we did not assay facial expression of this gene. (G and N) npnt expression is most intense along the lateral edge of facial endoderm (asterisk), where the proposed Itga8-FPC interactions occur. All scale bars are 100 Ám, applicable to their respective columns. |
| Genes: | |
|---|---|
| Fish: | |
| Anatomical Terms: | |
| Stage Range: | Pec-fin to Protruding-mouth |
Reprinted from Developmental Biology, 416(1), Talbot, J.C., Nichols, J.T., Yan, Y.L., Leonard, I.F., BreMiller, R.A., Amacher, S.L., Postlethwait, J.H., Kimmel, C.B., Pharyngeal morphogenesis requires fras1-itga8- dependent epithelial-mesenchymal interaction, 136-48, Copyright (2016) with permission from Elsevier. Full text @ Dev. Biol.