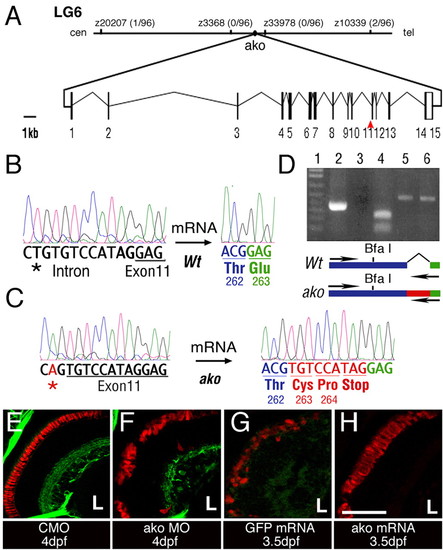
The ale oko locus encodes a zebrafish dynactin 2 homolog. (A) Map of the ako genomic region and exon/intron structure of the ako transcript. The position of mutant sequence in the akojj50 allele is indicated (red arrowhead). (B,C) Comparison of sequence trace data from wild-type (B) and mutant (C) animals. The akojj50 mutation activates a cryptic splice site in the genomic sequence. This, in turn, introduces a 9-bp insertion (red) that encodes a premature stop codon. (D) RT-PCR amplification of the dynactin 2 transcript using a primer that recognizes the mutant insertion sequence. Lane 1, DNA ladder; lane 2, amplification of the mutant transcript produces a single robust band; lane 3, amplification of the wild-type transcript does not produce a detectable product; lane 4, BfaI digestion confirms the identity of the amplification product from the mutant; lanes 5 and 6, control PCR amplification of actin from wild-type (5) and mutant (6) animals. The positions of amplification primers in the wild type and mutant are indicated on the diagram below. (E-H) Phenocopy and rescue of photoreceptor defects in ako mutant animals. Transverse cryosections through embryonic retinae were stained with the Zpr-1 antibody to visualize double cones (red). Green signal in E,F originates from the Tg(brn3c:mGFP) transgene. The injection of ako (H), but not GFP (G), mRNA rescues photoreceptor morphology in ako mutants. L, lens. Scale bar: 40 μm.
|

