- Title
-
The miR-144/Hmgn2 regulatory axis orchestrates chromatin organization during erythropoiesis
- Authors
- Kretov, D.A., Folkes, L., Mora-Martin, A., Walawalkar, I.A., Imrat, ., Syedah, N., Vanuytsel, K., Moxon, S., Murphy, G.J., Cifuentes, D.
- Source
- Full text @ Nat. Commun.
|
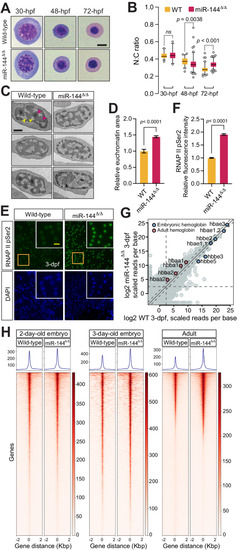
Loss of miR-144 impairs chromatin condensation during erythropoiesis. |
|
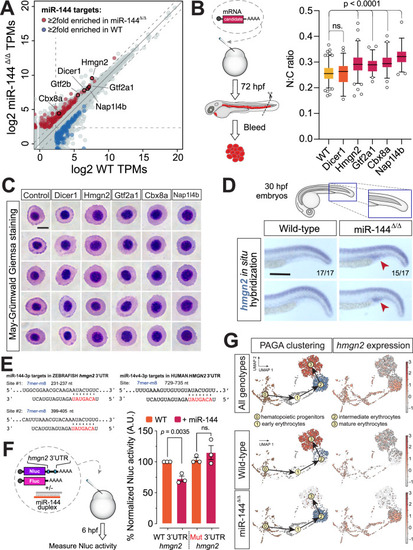
Hmgn2 is a miR-144 target whose expression is downregulated in mature erythrocytes. |
|
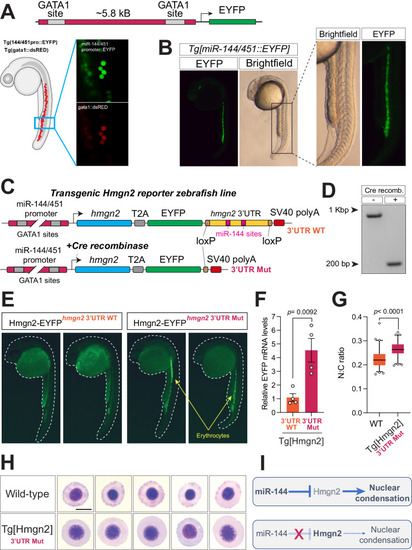
Disruption of miR-144-mediated repression of Hmgn2 impairs erythropoiesis. |
|
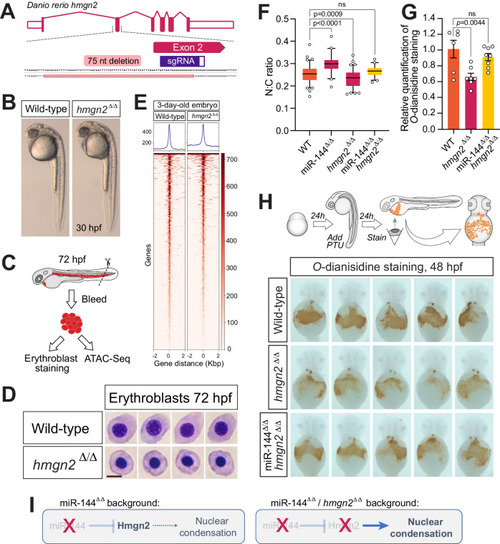
Reduction of Hmgn2 activity rescues the loss of miR-144 in erythropoiesis. |
|
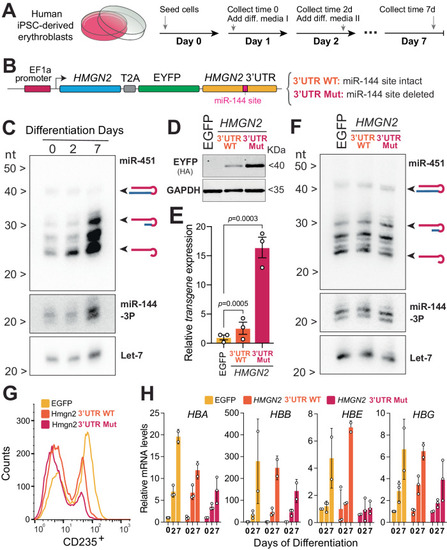
Probing the HMGN2/miR-144 regulatory axis in human erythroid progenitor cells. |