- Title
-
Core planar cell polarity genes VANGL1 and VANGL2 in predisposition to congenital vertebral malformations
- Authors
- Feng, X., Ye, Y., Zhang, J., Zhang, Y., Zhao, S., Mak, J.C.W., Otomo, N., Zhao, Z., Niu, Y., Yonezawa, Y., Li, G., Lin, M., Li, X., Cheung, P.W.H., Xu, K., Takeda, K., Wang, S., Xie, J., Kotani, T., Choi, V.N.T., Song, Y.Q., Yang, Y., Luk, K.D.K., Lee, K.S., Li, Z., Li, P.S., Leung, C.Y.H., Lin, X., Wang, X., Qiu, G., DISCO (Deciphering disorders Involving Scoliosis and COmorbidities) study group, Watanabe, K., Japanese Early Onset Scoliosis Research Group, Wu, Z., Posey, J.E., Ikegawa, S., Lupski, J.R., Cheung, J.P.Y., Zhang, T.J., Gao, B., Wu, N.
- Source
- Full text @ Proc. Natl. Acad. Sci. USA
|
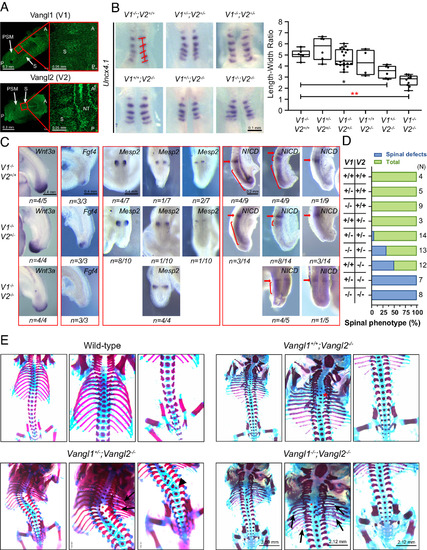
Somite development defects and vertebral malformations in Vangl1 and Vangl2 mutant mouse embryos. (A) Vangl1 and Vangl2 expression in the PSM, NT, and somites (S) of E8.5 wild-type embryos detected by immunofluorescence of endogenous Vangl proteins. A, anterior; P, posterior. (B) Whole-mount in situ hybridizations of Uncx4.1 in 5 to 6 somites stage of mouse embryos. ImageJ quantified the width and length of somites. The width was calculated on the basis of the average of each somite and the length was determined by the Uncx4.1 positive region along the A?P body axis. The significance of the differences between the groups was calculated by a one-way ANOVA test, F = 16.18, *P = 0.0038, **P < 0.0001. Box plots show the center line as the median, box limits as the upper and lower quartiles, and whiskers as the minimum to maximum values. (C) Whole-mount in situ hybridizations of Wnt3a, Fgf4, and Mesp2 and whole-mount immunohistochemistry of NICD in 23 to 25 somite stage of Vangl mutant mice at E9.5. The red arrow and bracket denote a variable cyclic pattern. (D) Penetrance of spinal phenotypes (rib and vertebrae) of Vangl mutant mice. The number of analyzed animals for each genotype is listed on the right side of the chart. (E) Spinal phenotypes of E17.5 to E18.5 Vangl1 and Vangl2 mutant mice. Red asterisks indicate vertebral fusion, vertebral misalignment, and delayed vertebral ossification. Arrowheads point to hemivertebrae, and arrows point to rib fusion, rib shortening/missing, and rib bifurcation. V1 denotes Vangl1, and V2 represents Vangl2. |
|
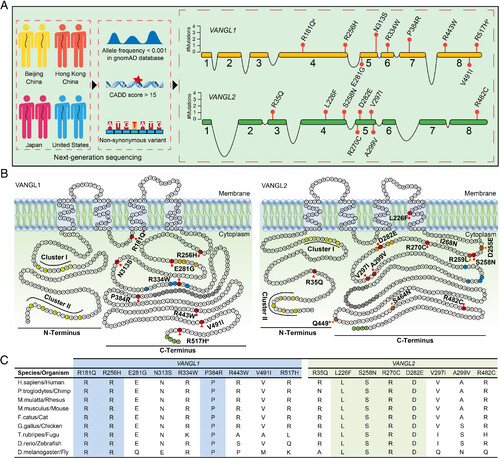
Identification of VANGL1 and VANGL2 variants in CS patients. (A) Brief workflow of the identification of VANGL1 and VANGL2 variants in a multicenter CS cohort (Left) and the location of the identified variants (Right). The numbered orange and green boxes indicate the coding exons of VANGL1 and VANGL2. (B) Amino acid residues of VANGL1 and VANGL2 defining sequence landmarks and signature motifs are depicted in different colors, including rare variants identified in our multicenter cohort (red), looptail variants (dark orange), Curly Bob variant (pink), two highly conserved phosphorylation clusters (yellow, cluster I and cluster II), trans-Golgi network sorting motif (orange), p97/VCP-interacting motif (blue), coiled-coil domain (gray), and PDZ-binding motif (light green). aThe same variant was found in reported NTDs patients. (C) Conservation and multiple alignments of VANGL1 and VANGL2 variants. Highlighted variants are conserved among all listed species. |
|
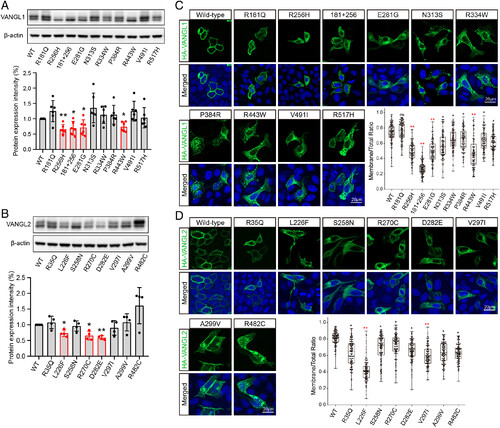
Effects of VANGL1 and VANGL2 variants on expression and localization. (A and B) Protein expression of VANGL1 and VANGL2 mutants in HEK293T cells. Band intensities of western blot gels were quantitated using ImageJ and normalized with ?-actin. Each experiment was repeated at least three times. The significance of the differences was calculated by a one-way ANOVA test, F = 8.77 (A), 10.67 (B). Error bars show the mean ± SD. The panels labeled in red show a significant difference, compared with the corresponding wild-type group, *P < 0.05, **P < 0.01. (C and D) Intracellular localization of wild-type and mutant VANGL1 and VANGL2 proteins. The HA-tagged VANGL1/2 proteins were stained in green, and nuclei were visualized by DAPI in blue. Membrane localization ratios of wild-type and mutant VANGL1 and VANGL2 were analyzed by ImageJ in more than 100 cells for each group. The significance of the differences was calculated by a one-way ANOVA test, F = 114.50 (C), 69.72 (D). *P < 0.05, **P < 0.01, and the mean difference between wild-type and labeled groups exceeded 0.2. Box plots show the center line as the median, box limits as the upper and lower quartiles, and whiskers as the minimum to maximum values. |
|
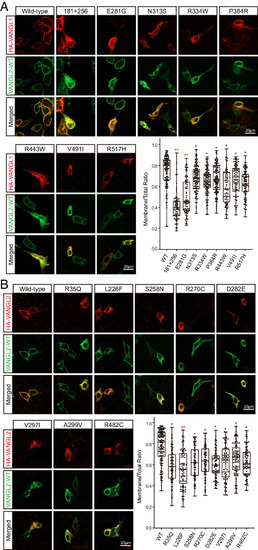
VANGL1 and VANGL2 variants exhibit dominant-negative effects. (A and B) Colocalization of HA-tagged wild-type and mutant VANGL1/2 (red) with Flag-tagged wild-type VANGL2 (green) in MDCK cells. Membrane localization ratios of wild-type and mutant VANGL1 and VANGL2 were analyzed by ImageJ in more than 100 cells for each group. The significance of the differences was calculated by a one-way ANOVA test, F = 49.04 (A), 17.24 (B). *P < 0.05, **P < 0.01, and the mean difference between wild-type and labeled groups exceeded 0.2. Box plots show the center line as the median, box limits as the upper and lower quartiles, and whiskers as the minimum to maximum values. |
|
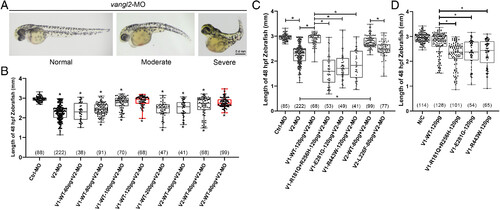
Mutant VANGL1 and VANGL2 fail to rescue CE defects in zebrafish embryos. (A) Illustrations of normal, moderate, and severe defects in CE in zebrafish embryos injected with vangl2-MO (morpholino). (B) The optimal dosage of human VANGL1 and VANGL2 wild-type mRNA to rescue CE defects in zebrafish embryos. The reduced body length of zebrafish embryos caused by 8 ng vangl2-MO was fully rescued by 120 pg human VANGL1 or 80 pg human VANGL2 wild-type mRNA. The significance of the differences was calculated by a one-way ANOVA test, F = 47.60. The Top and Bottom asterisks indicate significant differences compared to the control-MO group and the vangl2-MO group, respectively. (C) The comparison of body length in zebrafish embryos between control-MO (Ctrl-MO), vangl2-MO (V2-MO), and rescue groups. The significance of the differences was calculated by a one-way ANOVA test, F = 117.60. (D) The comparison of body length in zebrafish embryos after mRNA injection between blank control, human VANGL1 wild-type, and human VANGL1 mutant groups. The statistical significance of differences between the groups was calculated by the one-way ANOVA test, F = 41.83. *P < 0.05. Box plots show the center line as the median, box limits as the upper and lower quartiles, and whiskers as the minimum to maximum values. The number in parentheses indicates the number of analyzed zebrafish embryos. V1 and V2 denote Vangl1 and Vangl2, respectively. PHENOTYPE:
|
|
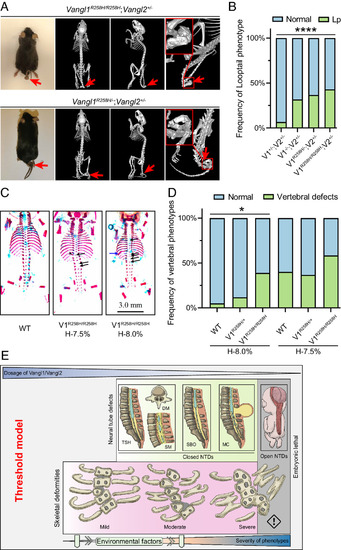
Vangl1-R258H knock-in mice display vertebral defects in a Vangl gene dose- and hypoxia-dependent manner. (A) Phenotype of the 2-mo-old Vangl1R258H/R258H;Vangl2+/? and Vangl1R258H/?;Vangl2+/? mice by MicroCT. Looptail was observed, and vertebral malformations are indicated by red arrows. (B) Penetrance of looptail (Lp) in various Vangl1-R258H mutant mice. The significance of the differences between the groups was calculated by a chi-squared test, ?2 = 27.56, P < 0.0001. (C) Dorsal view of skeletal preparation of E17.5 Vangl1R258H/R258H embryos with hypoxia. Black arrows indicate vertebral ossification abnormalities; blue arrows indicate fused rib; arrowheads indicate hemivertebrae. (D) The penetrance of vertebral defects in wild-type, Vangl1R258H/+, and Vangl1R258H/R258H mouse embryos induced by mild hypoxia. The significance of the differences between the groups was calculated by the chi-squared test, ?2 = 8.00 (H-8.0%), 2.77 (H-7.5%). *P < 0.05. V1 and V2 denote Vangl1 and Vangl2, respectively. H-8.0% and H-7.5% denote hypoxia at 8.0% and 7.5% oxygen concentration, respectively. (E) Threshold model of Vangl gene dosage in birth defects. The decrease in Vangl1 and Vangl2 gene dosage and the addition of environmental factors increase the penetrance and severity of skeletal and neurologic phenotypes. The threshold of Vangl gene dosage for vertebral defects and NT defects is different, with the former more sensitive to Vangl dosage. TSH, tethered spinal cord; DM, diastematomyelia; SM, syringomyelia; SBO, spinal bifida occulta; MC, meningocele; NTDs, neural tube defects. |