- Title
-
Lactylation-driven FTO targets CDK2 to aggravate microvascular anomalies in diabetic retinopathy
- Authors
- Chen, X., Wang, Y., Wang, J.N., Zhang, Y.C., Zhang, Y.R., Sun, R.X., Qin, B., Dai, Y.X., Zhu, H.J., Zhao, J.X., Zhang, W.W., Ji, J.D., Yuan, S.T., Shen, Q.D., Liu, Q.H.
- Source
- Full text @ EMBO Mol. Med.
|
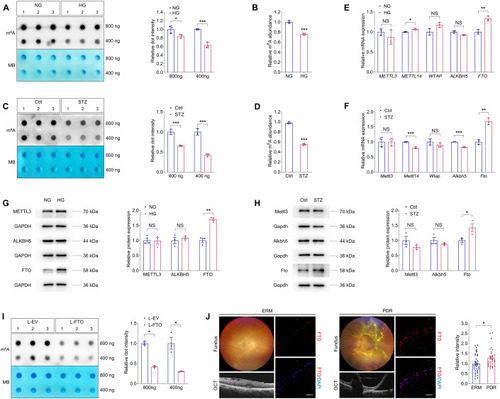
Identification of FTO as a potential regulator of DR ( |
|
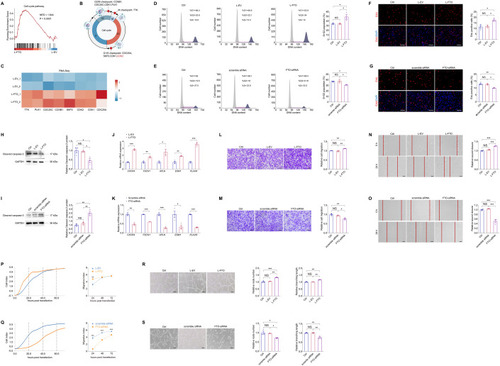
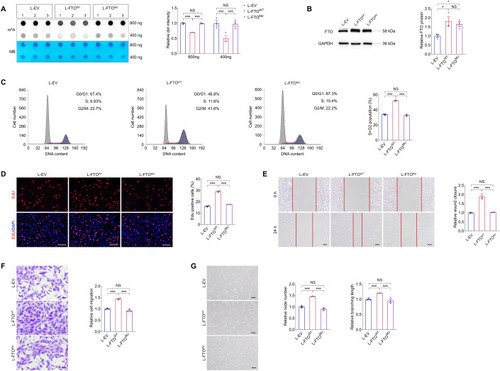
FTO promotes endothelial cell cycle progression and tip cell formation to facilitate angiogenesis in vitro. ( |
|
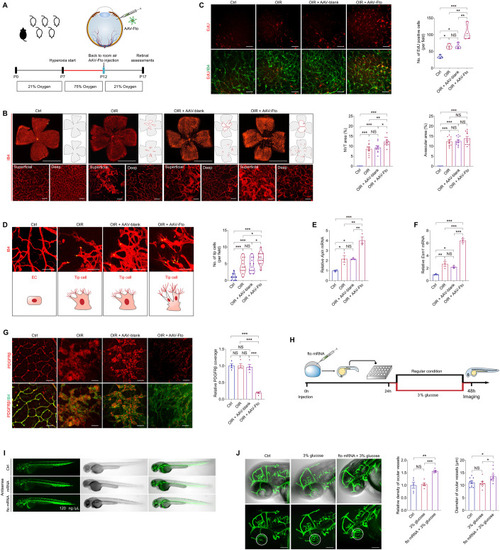
FTO promotes endothelial tip cell formation to facilitate neovascularization in mice and zebrafish. ( EXPRESSION / LABELING:
|
|
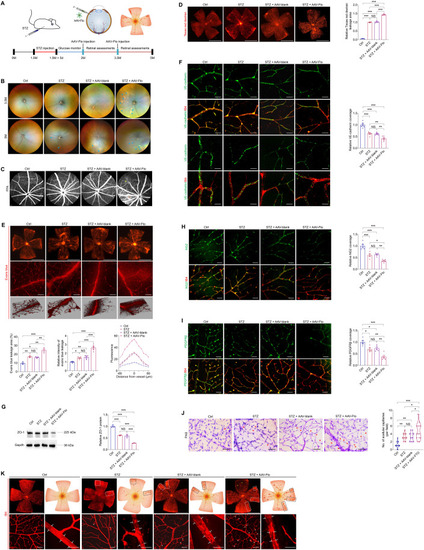
FTO regulates EC-pericyte crosstalk to trigger diabetes-induced microvascular dysfunction in mice. ( |
|
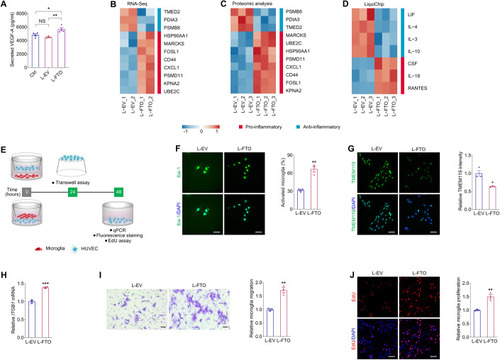
FTO triggers vascular inflammation and regulates EC-microglia crosstalk in vitro. ( |
|
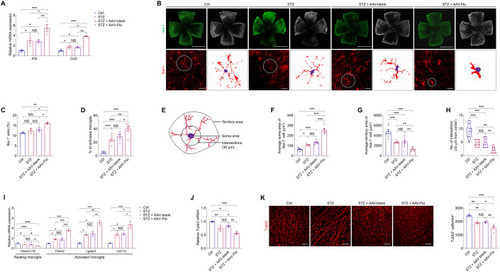
FTO triggers microglia activation and neurodegeneration in mice. ( |
|
Demethylation activity is required for FTO to regulate EC features ( |
|
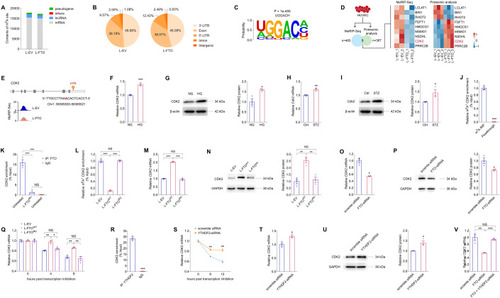
FTO regulates CDK2 mRNA stability with YTHDF2 as the reader in an m6A-dependent manner. ( |
|
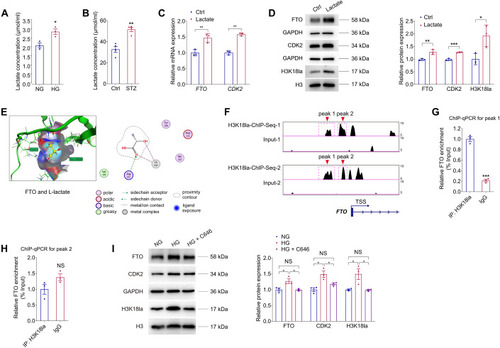
Lactic acid regulates FTO expression via histone lactylation. ( |
|
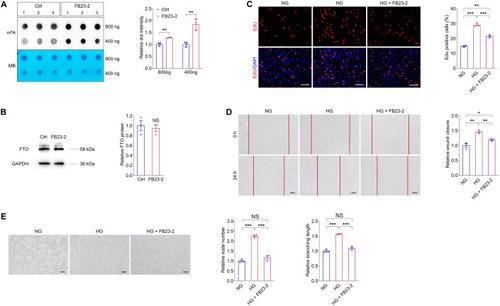
FB23-2 suppresses demethylation activity of FTO to inhibit diabetes induced endothelial phenotypes in vitro. ( |
|
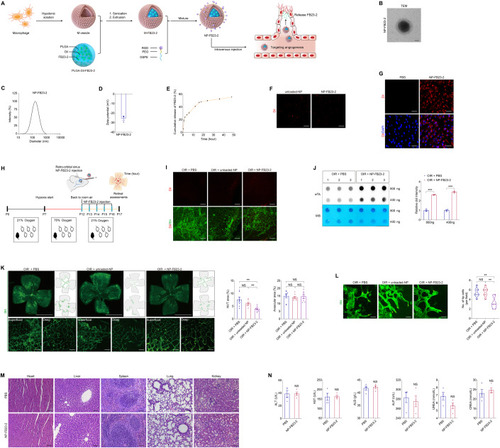
Characteristics of NP-FB23-2 and evaluation of its therapeutic efficacy on retinal neovascularization in mice. ( |
|
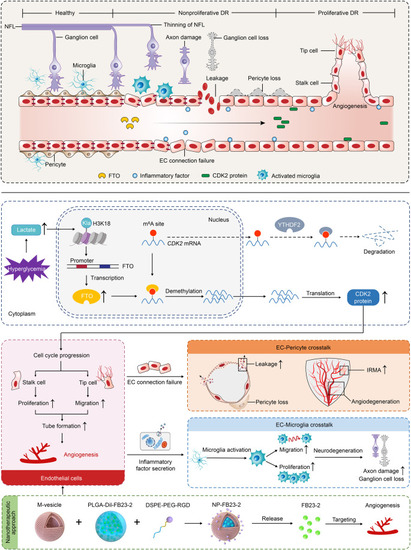
Schematic diagram of FTO-mediated effects on DR. Driven by histone lactylation H3K18la, FTO up-regulation in ECs triggers vascular endothelial tip cell formation and its crosstalk with pericyte and microglia to aggravate diabetes-induced microvascular dysfunction. FTO mediates DR phenotypes via regulating |