- Title
-
Notch signaling regulates cardiomyocyte proliferation during zebrafish heart regeneration
- Authors
- Zhao, L., Borikova, A.L., Ben-Yair, R., Guner-Ataman, B., MacRae, C.A., Lee, R.T., Burns, C.G., and Burns, C.E.
- Source
- Full text @ Proc. Natl. Acad. Sci. USA
|
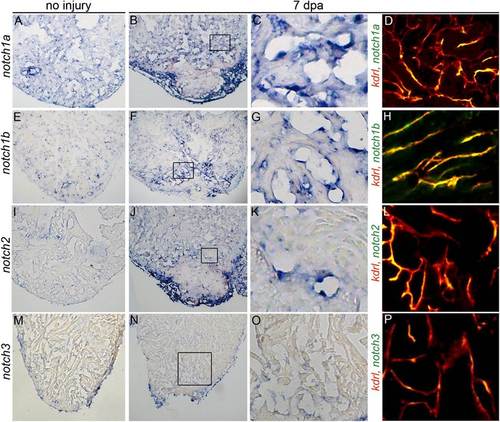
Ventricular apex amputation induces up-regulation of Notch receptor expression in endocardial and epicardial cells. Representative histological sections of uninjured (A, E, I, and M) and injured hearts (B, F, J, and N) at 7 d postamputation (dpa) showing up-regulation of three Notch receptors in the endocardium and/or epicardium following amputation injury by colormetric in situ hybridization analyses. Boxed regions of B, F, J, and N are shown at higher magnifications in C, G, K, and O. (D, H, L, and P) Representative cardiac sections from Tg(kdrl:mCherry) zebrafish processed for fluorescent in situ hybridization to visualize expression of the indicated Notch receptors and mCherry immunohistochemistry to visualize endocardial cells. Notch1a, notch1b, notch2, and notch3 transcripts colocalized with endocardial cells without expression in the intervening myocardium. (M and N) Notch3 transcripts were unchanged by injury. Consistent staining patterns were observed in eight hearts per experimental group for the colorimetric analyses and in two hearts for the fluorescent analyses. |
|
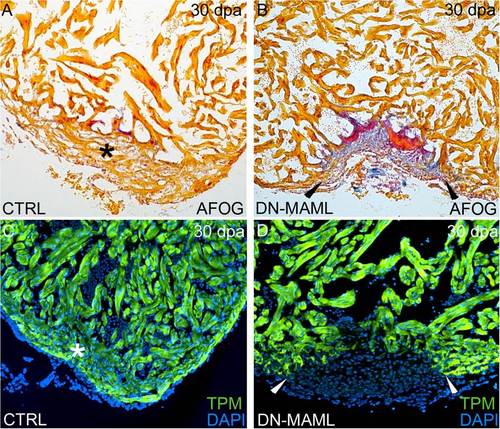
Inhibition of Notch signaling following amputation injury prevents cardiac regeneration. (A?D) Inhibition of Notch signaling impairs cardiac regeneration. Representative cardiac sections from heat shocked control (A and C) and Tg(hsp70:DN-MAML) (B and D) animals 30 dpa evaluated by AFOG staining (A and B) or immunofluorescence (C and D) for the myocardial marker TPM. Whereas heat shocked control animals robustly regenerated myocardium (asterisks in A and C), Tg(hsp70:DN-MAML) hearts failed to regenerate new myocardium (arrowheads in B and D) instead showing evidence of residual fibrin (red in B) and collagen deposition (blue in B). Cardiac regeneration failed in 0 of 24 heat shocked control animals and 34 of 35 heat shocked Tg(hsp70:DN-MAML) animals. |
|
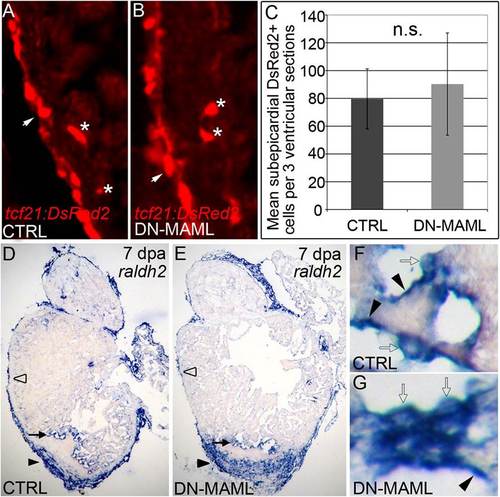
Notch signaling is dispensable for injury-induced epicardial and endocardial activation. (A and B) Representative cardiac sections from 7dpa heat shocked control (CTRL) (A) and Tg(hsp70:DN-MAML) (B) animals carrying the Tg(tcf21:DsRed2) transgene. In regions away from the wound, DsRed2+ epicardial cells (arrows) and EPDCs (asterisks) were grossly normal in Notch suppressed hearts. Seven of seven hearts in each experimental group expressed DsRed2 in epicardium and EPDCs. (C) Graph representing the average numbers of subepicardial DsRed2+ cells quantified in three ventricular sections from four hearts per experimental group. n.s., not significant. (D?G) Representative cardiac sections from heat shocked control (D) and Tg(hsp70:DN-MAML) (E) animals at 7 dpa stained for raldh2 transcripts by in situ hybridization. In both experimental groups, prominent raldh2 expression was observed in epicardial cells covering the wound (black arrowheads in D and E), away from the wound (open arrowheads in D and E), and in endocardial cells near the wound edge (black arrows in D and E). Higher magnifications of endocardial raldh2 expression are shown in F and G. Endocardial cells near the wound displayed both flattened (black arrowheads in F and G) and rounded morphologies (open arrows in F and G). Four of four hearts in each experimental group expressed endocardial and epicardial raldh2 transcripts. |
|
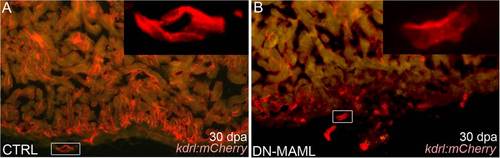
Evidence for regeneration of coronary endothelium in Notch-inhibited hearts. (A and B) Representative cardiac sections from heat shocked control (CTRL) (A) and Tg(hsp70:DN-MAML) (B) animals carrying the Tg(kdrl:mCherry) endothelial reporter at 30 dpa. (Insets) Higher magnification views of boxed regions in A and B. Whereas control animals regenerated mCherry+ endocardium and coronary vessels, Tg(hsp70:DN-MAML) hearts harbored mCherry+ endothelial tubes in the nonregenerated region of the heart. The myocardium is autofluorescent in A and B. Fourteen of 14 control and 11 of 11 Tg(hsp70:DN-MAML) hearts demonstrated these phenotypes. |
|
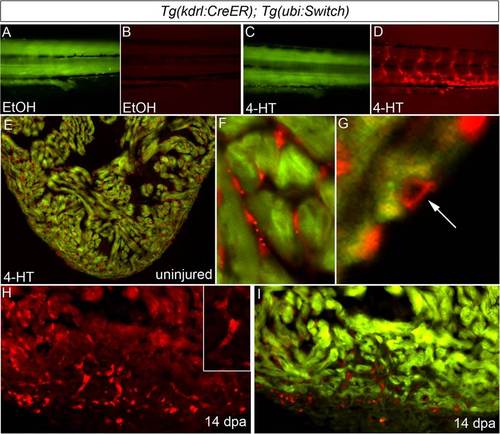
Regenerated coronary endothelium derives from preexisting endothelial cells during zebrafish heart regeneration. (A?D) Permanent labeling of endothelial cells during zebrafish embryogenesis. Tails of 96-hpf Tg(kdrl:CreER); Tg(ubi:Switch) embryos treated between 24 and 48 hpf with EtOH (A and B) or 4-HT (C and D) and imaged in the green (A and C) and red (B and D) channels. All 4-HT?treated embryos examined expressed mCherry in endothelial cells (n > 100), whereas EtOH-treated embryos failed to report red fluorescence (n > 100). (E?G) The adult endocardium and coronary endothelium displayed widespread and permanent mCherry labeling following 4-HT treatment during embryonic stages (n = 3/3). Representative cardiac section (E) and close up views of mCherry-labeled endocardium (F) and coronary endothelium (G). (H and I) Representative cardiac section from a regenerating Tg(kdrl:CreER); Tg(ubi:Switch) animal with mCherry-labeled endocardial and endothelial cells at 14 dpa. Numerous mCherry+ endothelial tubes (Inset) populated the regenerating myocardium. (n = 3/3.) |
|
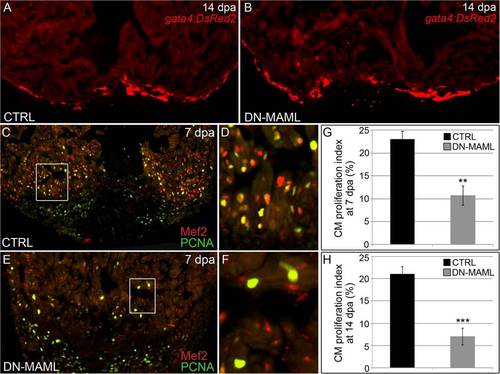
Notch signaling is required for cardiomyocyte proliferation during zebrafish heart regeneration. (A and B) Representative cardiac sections from heat shocked control (CTRL) (A) and Tg(hsp70:DN-MAML) (B) animals carrying the Tg(gata4:DsRed2) transgene at 14 dpa. DsRed2 expression was visible in the compact myocardium of both control and Tg(hsp70:DN-MAML) hearts (n = 12/12 in each group). (C?F) Representative cardiac sections from heat shocked control (C and D) and Tg(hsp70:DN-MAML) (E and F) animals at 7 dpa. Sections were double immunostained to identify cardiomyocyte nuclei (Mef2+) and nuclei undergoing DNA replication (PCNA+). Boxed regions in C and E are shown at higher zoom in D and F. The percentages of myocardial nuclei undergoing DNA replication near the wound edges were quantified at 7 (G) and 14 (H) dpa and reported as mean proliferation indices ± 1 SD, **P < 0.01 and ***P < 0.001. In the control group, seven and eight hearts were analyzed at 7 and 14 dpa, respectively. In the Tg(hsp70:DN-MAML) group, three and four hearts were analyzed at 7 and 14 dpa, respectively. |
|
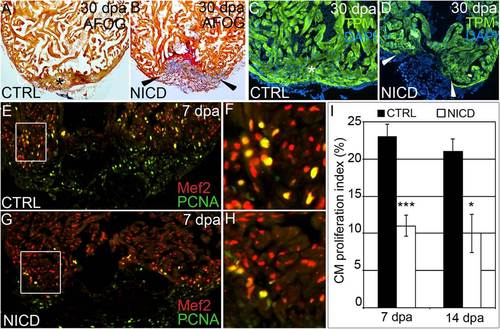
Hyperactivation of Notch signaling impairs cardiomyocyte proliferation and heart regeneration. (A?D) Representative cardiac sections from heat shocked control (CTRL) (A and C) and Tg(hsp70:Gal4); Tg(UAS:NICD) (B and D) animals at 30 dpa stained with AFOG (A and B) or an antibody recognizing the myocardial marker TPM (C and D). Whereas heat shocked CTRL animals regenerated myocardium (asterisks in A and C), Tg(hsp70:Gal4); Tg(UAS:NICD) hearts exhibited impaired regeneration (arrowheads in B and D), residual fibrin (red in B), and collagen deposition (blue in B). In the CTRL group, >25 hearts were analyzed and all showed significant myocardial regeneration. In the Tg(hsp70:Gal4); Tg(UAS:NICD) group, 18 hearts were analyzed and 14 showed a significant myocardial deficit, residual fibrin, and collagen deposition. (E?H) Representative cardiac sections of heat shocked CTRL (E and F) and Tg(hsp70:NICD) (G and H) animals at 7 dpa double stained with antibodies that recognize cardiomyocyte nuclei (Mef2+) and nuclei undergoing DNA replication (PCNA+). Boxed regions in E and G are shown at higher zoom in F and H. (I) The percentages of cardiomyocyte nuclei undergoing DNA replication were quantified along the wound edge at 7 and 14 dpa and expressed as mean proliferation indices ± 1 SD, ***P < 0.001; *P < 0.05. In the control group, seven and eight hearts were analyzed at 7 and 14 dpa, respectively. In the Tg(hsp70:NICD) group, three and four hearts were analyzed at 7 and 14 dpa, respectively. |
|
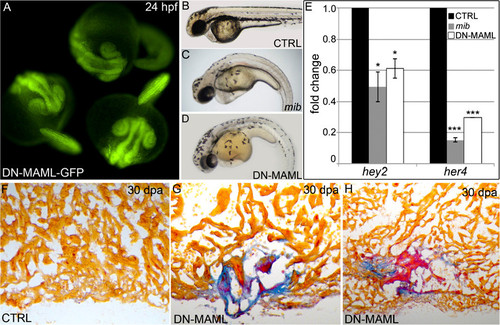
Heat shock inducibility of DN-MAML-GFP, suppression of Notch signaling by DN-MAML-GFP, and cardiac regenerative deficiencies in an independent Tg(hsp70:DN-MAML-GFP) line. (A) Embryos carrying the Tg(hsp70:DN-MAML-GFP)fb10 transgene were heat shocked for 30 min at 37 °C following gastrulation (bud stage). At 24 hpf, these embryos exhibited prominent GFP fluorescence indicating DN-MAML-GFP protein expression. (B?E) Heat-shock-inducible suppression of Notch signaling in Tg(hsp70:DN-MAML) animals. Control (CTRL) and Tg(hsp70:DN-MAML) embryos were heat shocked at bud stage (B and D) or 19 hpf (E) and analyzed for gross morphological defects at 48 hpf (B and D) or relative expression of Notch target genes hey2 and her4 by qPCR at 24 hpf (E). Mind bomb (mib) mutant embryos, which are devoid of Notch signaling, were analyzed in parallel. Whereas control embryos were unaffected by heat shock (B), mib (C), and heat shocked Tg(hsp70:DN-MAML) (D) embryos displayed similar body axis curvature defects. (E) Graph showing the relative expression levels of hey2 and her4 transcripts in each experimental group. Three biological replicates were included in the qPCR analysis. Data are presented as means ± 1 SD, *P < 0.05; ***P < 0.001. (F?H) AFOG staining of representative cardiac sections at 30 dpa from heat shocked control (CTRL; F) and transgenic adults carrying an independent insertion of the Tg(hsp70:DN-MAML-GFP) transgene [Tg(hsp70:DN-MAML-GFP)fb11] (G and H). Similar to Tg(hsp70:DN-MAML)fb10 hearts, heat shocked Tg(hsp70:DN-MAML)fb11 hearts displayed prominent regenerative deficiencies as evidenced by fibrin retention (red) and collagen deposition (blue). Sections shown in G and H are from independent animals. Cardiac regeneration failed in 0 of 8 heat shocked control hearts and 5 of 5 heat shocked DN-MAMLfb11 hearts. |
|
Dependence of mCherry reporter fluorescence on 4-HT exposure. (A?C) Representative cardiac section from an untreated (no exposure to 4-HT) Tg(kdrl:CreER); Tg(ubi:Switch) double transgenic adult zebrafish imaged in the green (A) and red (B) channels. Merged image shown in C. No mCherry reporter fluorescence was visible (n = 4). |
|
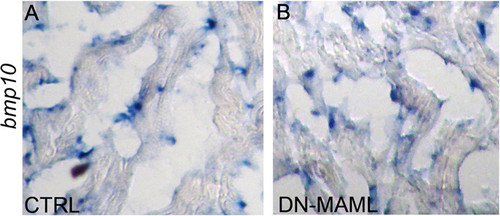
Bmp10 expression appears unchanged in Notch-suppressed hearts. (A and B) In situ hybridization showing bmp10 transcripts in cardiac sections from control and Notch-suppressed hearts at 7 dpa. In both cases, bmp10 transcripts localized to presumptive endocardial cells, and myocardial cells appeared devoid of bmp10 transcripts. |