- Title
-
ErbB2 and ErbB3 regulate amputation-induced proliferation and migration during vertebrate regeneration
- Authors
- Rojas-Muñoz, A., Rajadhyksha, S., Gilmour, D., van Bebber, F., Antos, C., Rodríguez Esteban, C., Nüsslein-Volhard, C., and Izpisúa Belmonte, J.C.
- Source
- Full text @ Dev. Biol.
|
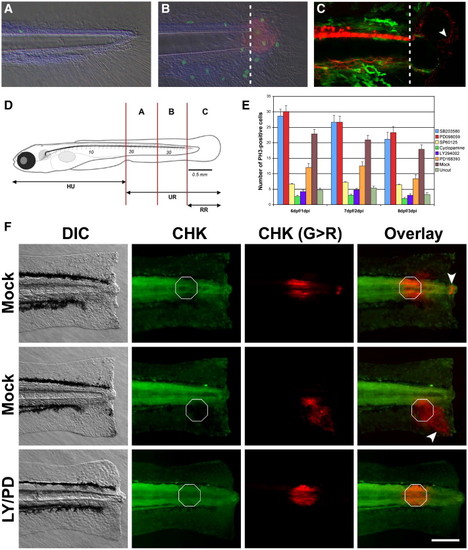
A chemical genetic screen in zebrafish identifies several oncogenes as regulators of epimorphic regeneration. (A) Zebrafish larvae are able to regenerate a significant portion of the tail that includes spinal cord, muscle, skin and blood vessels. At 6 dpf very few proliferating cells (visualized in green with anti-Phospho Histone H3 antibodies) are detected in the caudal region of control larvae. (B) Conversely, a marked increment of mitotic cells can be observed in amputated larvae at 6 dpf/1 dpi. Some, but not all of the dividing cells are present in the blastema (visualized in red with an antibody anti-Vimentin). (C) Transgenic lines NBT:DsR2 (van Drenth, M., unpublished) and FoxD3:EGFP indicate that the blastema is heavily innervated (white arrowhead, red channel) and void of neural crest cells at 1 dpi (green channel). Dotted lines indicate the amputation plane in panels (B) and (C). Amputation-induced proliferation of progenitor cells can be modulated by small chemical compounds. (D) In order to quantify the proliferative response, the number of dividing cells is counted in each one of the compartments depicted and (E) tabulated in the presence or absence of small compound inhibitors specific for a number of oncogenes. Data for the regions A + B (see panel D) is shown (E). At 6 dpf/1 dpi, the number of proliferating cells in region B is significantly enhanced by incubating regenerating larva in 20 μM SB203580 and 20 μM PD098059. In contrast, this number is significantly reduced in larva incubated in 20 μM SP600125, 5 μM Cyclopamine, 5 μM LY294002 or 10 μM PD168393. (F) Different cell types migrate towards the amputation plane during regeneration. Larvae were amputated at 5 dpf followed by immediate photoconversion of the fluorescent protein CoralHue? Kaede at localized spots (white octagons). At 24 hpi different cell types, including cells from the epidermis and the notochord (white arrowheads), move towards the amputation plane in larvae incubated in DMSO. Conversely, no cell movement was observed in larvae incubated in 10 μM PD168393/LY294002 during the same period of time. Importantly, cells from uncut larvae do not undergo migration over the time window of our experiments (data not shown). Under our conditions, UV light photoconverts many cells along the L/R axis at the given A/P position in the larvae. This is useful for the main reason that as most photoconverted cells do not move towards the amputation plane they clearly identify the initial A/P position of the photoconverted cluster at 24 hpi (as indicated by the white octagons in the panel). For the purpose of morphological reference: HU, head to urogenital region; UR, urogenital region; RR, regenerated region. Scale bar 100 μm. |
|
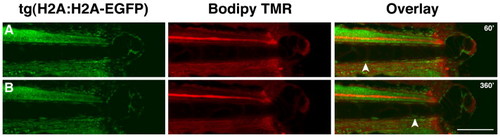
Cells from the notochord contribute to the regeneration blastema. Strip of images taken from two-color time-lapse of a transgenic tg(H2A:H2A-EGFP) larvae injected with Bodipy-TMR, with the time of image capture shown in minutes after amputation. Tracking of individual cells during this period of time indicates that cells from the notochord (white arrowheads) are rapidly deposited into the blastema (see also Movie S1). Scale bar 50 μm. |
|
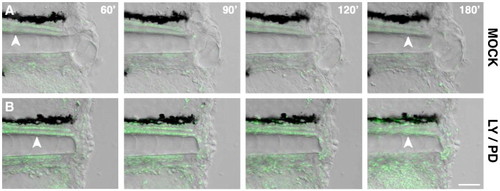
Simultaneous inhibition of EGFR and PI3K signaling affects regeneration. Transgenic animals tg(H2A:H2A-EGFP) incubated in 5 μM LY294002 and 10 μM PD168393 from the amputation moment onwards exhibit impaired localization of notochord cells and affect the timely formation of the regeneration blastema (B), compared to controls incubated in DMSO (A) (white arrowheads). The migration of highly motile cells types to the amputation plane is not affected in treated animals compared to controls (Movie S2 and S3). Scale bar 20 μm. |
|
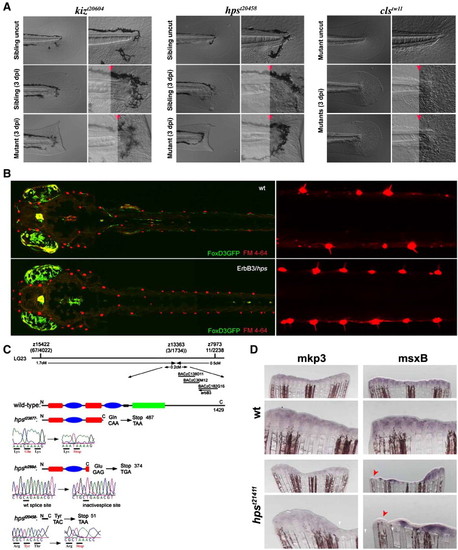
ErbB2 and ErbB3 are required for regeneration. (A) Two zygotic recessive regeneration mutants, kitzelig and hypersensitive, have been identified from a bench mutagenesis screen performed in zebrafish larva. DIC images clearly show that the caudal fin does not regenerate upon amputation on the homozygous mutants after 3 days post-injury (3 dpi). The amputation plane can be identified as the point where the notochord ends abruptly. In the middle and lower panels the amputation plane is marked with a red arrowhead. In contrast, homozygous mutants for a null allele of sox10/colorless do regenerate. The right hand panels are larger magnifications of the left panels. (B) kitzelig and hypersensitive carry mutations on the EGFR members ErbB2 and ErbB3, respectively. FM 4-64 staining of neuromasts (red) of hps larvae at 7 dpf shows strongly increased number of neuromast when compared to wild-type siblings (dorsal view). Both fish carry a FoxD3GFP transgene that labels neural crest derivatives (green). To the right a close up dorsal view of trunk shows neuromast hair cells protruding from every myotome in hps larvae. (C) Mapping placed hps on LG23 with tight linkage to a recently characterized erbB3 homologue. Sequencing confirmed the presence of nonsense mutations in three alleles (plots) and block-diagram shows the resultant truncated proteins (domains: Cysteine-Rich (red), Kinase (green), Furin (blue), Transmembrane (black)). (D) Expression of msxB is affected in erb3 mutants at 3 dpi. Adult fish homozygous for the hpst21411 allele exhibit a normal pattern of mkp3 expression compared to siblings despite of significant morphological defects on their regenerate. In contrast, the expression of msxB is downregulated in localized areas of homozygous mutants (red arrowheads) compared to controls. These areas exhibit a marked reduction of proliferating progenitor cells as defined by the number of Phospho-HistoneH3 positive cells (see Fig. 8A). EXPRESSION / LABELING:
PHENOTYPE:
|
|
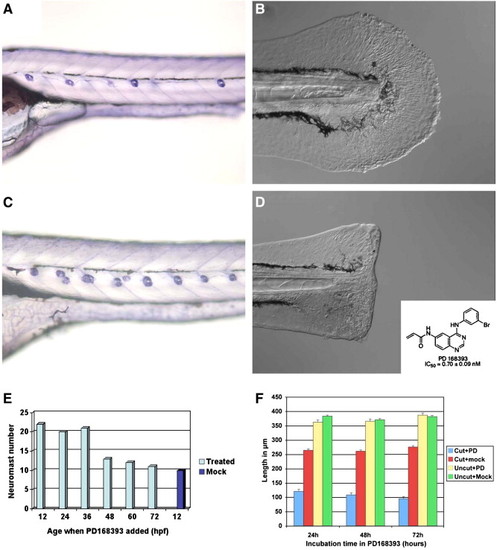
The ErbB inhibitors PD168393 and AG1478 phenocopy the mutant phenotypes observed in kiz and hps. (A and B) Compared to Mock incubated controls the number of neuromasts in the lateral line system doubles (C) and the regenerative response is blocked (D) upon incubation with either 10 μM PD168393 or AG1478. (E) The function of ErbB receptors is needed before 36 hpf in the lateral line system and its inhibition blocks growth post-injury during the regeneration response in the caudal fin even when added at 48 hpi (F). The data is expressed as Incubation time in the inhibitor (h)/time when inhibitor was added (hpi). As the inhibitor irreversibly binds to ErbB the data corresponds to 24h/48, 48h/24 hpi and 72h/0hpi (F). RR length (see Fig. 1D) was digitally measured at 8 dpf/3 dpi using the Scaling option in AxioVision 4.4. Anterior to the left, dorsal up. |
|
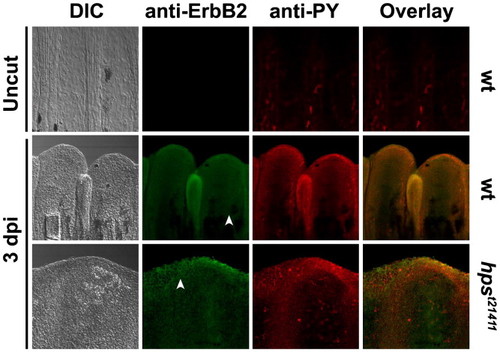
ErbB2 is expressed upon amputation all along the regenerating caudal fin of adult fish. Before amputation (Uncut) the amount of ErbB2 in the caudal fin is undetectable. However, ErbB2 is strongly accumulated after 1 dpi (Fig. S5) and 3 dpi. Similarly, the degree of protein phosphorylation at Y residues is low before amputation compared to amputated wild-type fins at 1 dpi and 3 dpi. Homozygous adults for hpst21411 exhibit a similar pattern of ErbB2 expression and Y-phosphorylation compared to wild-type fins, despite the absence of regeneration. Like in larval stages, ErbB2 is present all over the fin, including the plasma membrane of blastema cells (white arrowheads) and the neuromasts. In all cases, the ErbB2 signal often overlaps with the phospho-Y signal. EXPRESSION / LABELING:
|
|
Initial events of wound healing occur in ErbB3 mutants. (A and B) In wild-type adults, caudal fin regeneration is completed around 10 days post-injury (dpi). (C and C′) Conversely, homozygous mutants for the ErbB3 allele hpst21411 do not regenerate properly even after 30 dpi. (D) At the histological level, closure of the wound epidermis is not affected in homozygous mutants for hpst21411 at 1 dpi compared to wild-type controls although the formation of the blastema is delayed. At 3 dpi, the blastema in homozygous mutants is smaller compared to wild-type controls. Additionally, the distal blastema is absent in some individuals (arrowhead on inset). All sections are longitudinal. Scale bar 50 μm. (E) Polymorphonuclear leukocytes (PMNs) migrate to the wound site in wt animals, ErbB3 mutants and wt animals incubated with ErbB/PI3K (PD/LY) inhibitors. The initial site of DiI injection and the location of putative PMNs at 24 hpi is demarcated with white and red arrowheads, respectively. Most labeled cells migrate towards the amputation plane. PHENOTYPE:
|
|
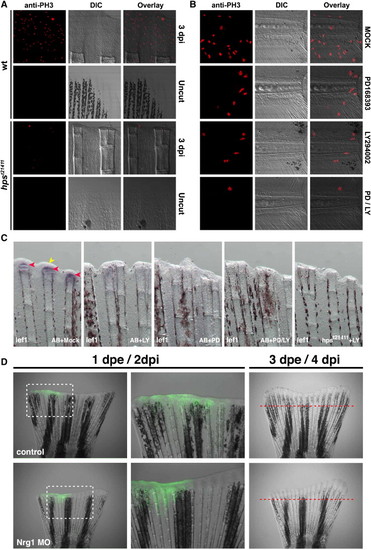
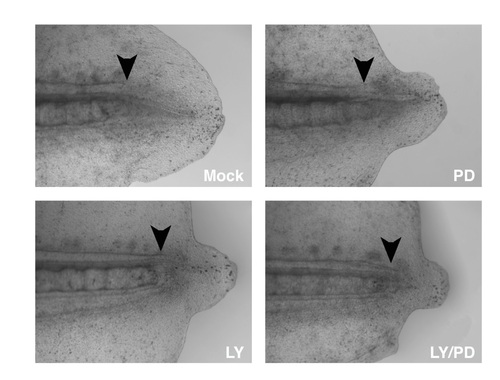
PI3K functionally interacts with ErbB2 and ErbB3 during regeneration. (A) Adult homozygous mutants for hpst21411 have a significantly reduced amputation-induced progenitor proliferation measured after 3 dpi compared to wild-type controls (Fig. S6). Similarly, amputation-induced progenitor proliferation in the adult caudal fin at 1 dpi is reduced upon incubation with 5 μM of the PI3K inhibitor LY294002 or 10 μM of the ErbBs inhibitor PD168393 for 24h. Importantly, upon simultaneous incubation with both inhibitors the amount of proliferating cells is reduced even more than in fins incubated with either alone (data not shown). (B) Similar to the adult situation, the simultaneous incubation with the and ErbB inhibitors significantly diminished the number of proliferating cells in the region A + B of the regenerate compared to amputated larvae incubated with either inhibitor alone or in DMSO. This trend is observed all over the duration of the regeneration response (Fig. S6). (C) ErbB and PI3K signaling interact to modulate regeneration in zebrafish. Images of amputated caudal fins at 24 hpi in wild-type or hpst21411 ErbB3 homozygous mutants. Normally, lef1 is transcribed in the basal layer of the regenerating epidermis at 24 hpi (red arrowheads). The wounded epithelium is also clearly visible at this stage (yellow arrowhead). Animals incubated with 10 μM of the PI3K inhibitor LY294002 exhibit an evident decreased on lef1 expression. Incubation with 10 μM PD168393 show a phenotype similar to that observed with LY294002. The thickness of the wounded epithelium is markedly reduced upon incubation with 10 μM LY294002 and 10 μM PD168393. Similarly, homozygous mutants for the ErbB3 allele hpst21411 incubated with 10 μM LY294002 lack any detectable lef1 expression or wound epithelium. (D) Electroporation of a morpholino designed against NRG1 disturbs regeneration while a control morpholino designed against rx3 does not. The difference in length of the regenerates that grew within 3 days post electroporation (dpe) between the two treatments was significant (98% ± 5.3% for the control morpholino and 54.2% ± 7.1% for the Nrg1 MO, P <0.005). The data depicts the percent length of the regenerate of the experimental side in relation to the control side (e.g. the other half of the same fin). |
|
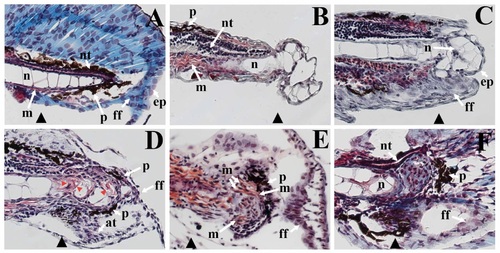
Regenerated structures in zebrafish larvae. (A) Masson?s trichrome staining of the paraffin embedded sections of caudal fins show the cells of the neural tube (nt), notochord (n), muscle fibers (m), as well as pigment cells (p) and epithelium (ep) of the finfold (ff) of an unamputated larval fin. The site of the prospective amputation plane is indicated with a black triangle. (B) 6 hours post injury (hpi), a protrusion from the amputation plane contains vacuolated cells containing very little cytoplasm, a cellular morphology like that of the cells of the notochord. (C) Between 12 and 24 hpi, an epidermal layer (ep), approximately two cells thick, covers the notochord protrusion. Also, finfold tissue (ff) has started to grow distally from the amputation stump. (D) By 3 day post injury (dpi), cells of the finfold now surround the protrusion, and actinotrichia (at, small skeletal filaments of the finfold) are present. Furthermore, cells within the protrusion no longer have the same morphology as cells from the notochord in that they contain more cytoplasm (red staining indicated by red arrowheads). Pigment cells (p, black melanophores) are now present dorsally and ventrally around the protrusion. (E) By 4 dpi, striated muscle fibers surround the protrusion of the fin regenerate. Melanophores (p) now encircle the protrusion, and the architecture of the finfold is very similar to that of the uninjured larval finfold. (F) At 7 dpi, the neural epithelium of the neural tube (nt) is now present in the fin regenerate. Although the basic architecture of the caudal fin is complete, tissue remodeling still appears to be ongoing: there still is a cluster of cells at the distal end of the notochord that have not integrated as a differentiated caudal fin tissue. |
|
Effect of small compounds on zebrafish larvae regeneration. Proliferating cells scored by anti-PH3 staining at 6 dpf / 1dpi. The number of proliferating cells is significantly enhanced by incubating regenerating larva in 20 μM SB203580 and 20 μM PD098059. In contrast, this number is significantly reduced in larva incubated in 20 μM SP600125, 5 μM Cyclopamine, 5 μM LY294002 or 10 μM PD168393. The images correspond to the region B as defined in Fig. 1D. |
|
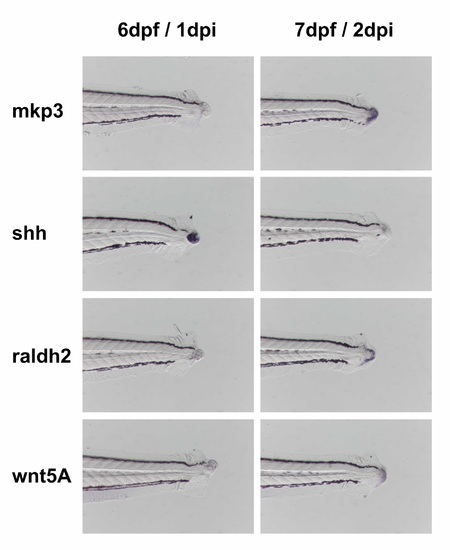
Expression of different signaling pathways components during larvae regeneration. Upon tail amputation zebrafish larvae exhibit distinct and dynamic expression patterns on transcripts usually upregulated during adult fin regeneration. The transcripts shown are components of four signaling pathways known to be involved in different contexts of regeneration (e.g. Hh, Wnt, RA and FGF). |

Expression of members of the NRG1 pathway during regeneration. The expression of erbB2, erbB3, and NRG1 is markedly increased after caudal fin amputation compared to controls uncut. Their expression is widely distributed over the length of the amputated fins including the blastema. |

ErbB2 is translated upon amputation in the regeneration blastema. Antibodies anti-ErbB2 (green panels) and anti-Phospho Y (red panels) are colocalized in the blastema and the Neuromasts of regenerating larvae. Single confocal sections suggest that ErbB2 is mainly localized to the cellular membrane of blastema cells (paraxial plane). Larvae incubated in 5 μM LY294002 and 10 μM PD168393 exhibit a delay in the formation of the blastema (PD/LY) but ErbB2 is still detected at the amputation plane (white arrowheads). Anterior to the left, dorsal up and 6dpf / 1dpi larvae in all panels. Scale bar 20 μm. |
|
The interaction between ErbB and PI3K in the context of regeneration is evolutionary conserved. The size and morphology of the regenerate is abnormal compared to controls upon incubating Axolotl larvae for 12 dpi in 10 μM PD168393, 10 μM LY294002 or a combination of the two inhibitors. The black arrowheads indicate the amputation plane. |
|
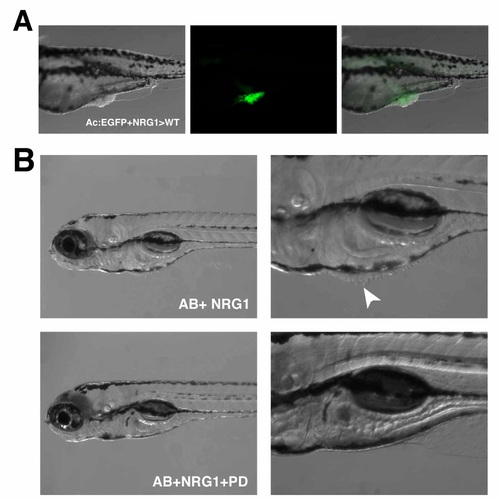
Neuregulin 1 induces proliferation through ErbB-PI3K. (A) Cells from tg(Actin:EGFP) embryos injected with mRNA for Neuregulin 1 (NRG1) at the one cell-stage were transplanted into wild type embryos at the 1K cellstage. Abnormal proliferation of both labeled and unlabeled cells were frequently observed when clones were present in the skin. (B) Wild-type animals injected with mRNA for NRG1 at the 1 cell-stage and incubated in the ErbB inhibitor PD168393 never developed abnormally proliferating cells compared to injected animals incubated in DMSO (white arrowhead). |
Reprinted from Developmental Biology, 327(1), Rojas-Muñoz, A., Rajadhyksha, S., Gilmour, D., van Bebber, F., Antos, C., Rodríguez Esteban, C., Nüsslein-Volhard, C., and Izpisúa Belmonte, J.C., ErbB2 and ErbB3 regulate amputation-induced proliferation and migration during vertebrate regeneration, 177-190, Copyright (2009) with permission from Elsevier. Full text @ Dev. Biol.