- Title
-
Fgf4 is required for left-right patterning of visceral organs in zebrafish
- Authors
- Yamauchi, H., Miyakawa, N., Miyake, A., and Itoh, N.
- Source
- Full text @ Dev. Biol.
|
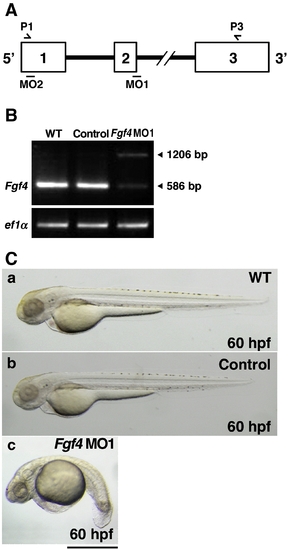
Inhibition of Fgf4 functions in zebrafish embryos. (A) The coding region of zebrafish Fgf4 is divided by two introns. Open boxes and black lines indicate exons and introns, respectively. MO1 and MO2 indicate the target positions of Fgf4 MO1 and MO2, respectively. P1 and P3 indicate sites of P1 and P3 primers. (B) Zebrafish two-cell embryos were injected with Fgf4 MO1 (∼ 45 ng). The entire coding region of Fgf4 cDNA was amplified from wild-type or Fgf4 MO1-injected embryonic cDNA by RT-PCR using P1 and P3 primers. Arrowheads (586 bp and 1206 bp) indicate mature and unspliced Fgf4 cDNAs, respectively. Elongation factor 1-alpha (ef1α) cDNA was also amplified as a control. The cDNAs were analyzed by 1.2% agarose gel electrophoresis. After electrophoresis, the gel was stained with ethidium bromide. (C) Two-cell embryos were injected with standard control MO and Fgf4 MO1 (∼ 45 ng). Lateral views of the embryos at 60 hpf are shown. Control MO-injected embryos (97.2%, n = 36) (b) as well as the wild-type embryos (a) developed well. In contrast, Fgf4 MO-injected embryos showed bent trunks, pericardial edema and cerebral atrophy (67.1%, n = 73) (c). A scale bar = 500 μm. PHENOTYPE:
|
|
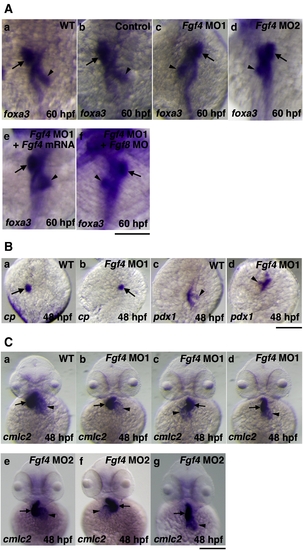
The effect of Fgf4 knockdown on left?right patterning. (A) The expression of foxa3, a marker gene for endoderm, was examined by whole mount in situ hybridization using a digoxigenin-labeled antisense foxa3 cRNA probe. The expression of foxa3 showed that the liver and pancreas were positioned to the left and right of the midline in wild-type embryos, respectively at 60 hpf (94.1%, n = 34) (a). Control MO-injected embryos also showed similar LR patterning of the liver and pancreas (93.8%, n = 32) (b). In contrast, the positions of these organs were randomized in Fgf4 MO1-injected embryos (normal (61.3%, n = 31); reversed (38.7%, n = 31)) (c). The expression of foxa3 showed similar randomized positioning in Fgf4 MO2-injected embryos (normal (56.7%, n = 30); reversed (43.3%, n = 30)) (d). The injection of Fgf4 mRNA significantly rescued the defects in Fgf4 MO1-injected embryos (80.6%, n = 31) (e). The injection of both Fgf4 MO1 and Fgf8 MO also revealed randomized positioning (58.1%, n = 31) (f). Arrows and arrowheads indicate the liver and pancreas, respectively. A scale bar = 200 μm. (B) The expression of ceruloplasmin and pdx1, marker genes for the liver and pancreas, respectively, in embryos at 48 hpf was examined. Their expression confirmed randomized positioning of the liver and pancreas (liver, normal (60%, n = 30); reversed (40%, n = 30): pancreas, normal (63.3%, n = 30); reversed (36.7%, n = 30)) in Fgf4 MO1-injected embryos (a?d). Arrows and arrowheads indicate the liver and pancreas, respectively. A scale bar = 200 μm. (C) The expression of cmlc2, a marker gene for the heart, in embryos at 48 hpf was examined. After the heart tube has formed, leftward movement of the heart (jogging) is followed by rightward bending of the ventricle (D-looping) in wild-type embryos (a). In contrast, Fgf4 MO1-injected embryos exhibited randomized looping (D-looping (38.5%, n = 78); reversed looping (L-looping) (47.4%, n = 78); no looping (14.1%, n = 78)) (b?d). Fgf4 MO2-injected embryos also exhibited randomized looping (D-looping (34.2%, n = 38); reversed looping (L-looping) (50%, n = 38); no looping (15.8%, n = 38)) (e?g). Arrows and arrowheads indicate the ventricle and atrium, respectively. A scale bar = 200 μm. |
|
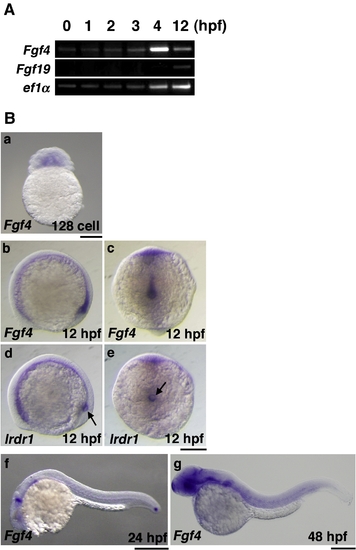
The expression of Fgf4 in zebrafish. (A) The expression of Fgf4 in embryos at 0?4 hpf was examined by RT-PCR. The expression of Fgf19 and ef1α was also examined as negative and positive controls, respectively. Fgf4 was expressed maternally. (B) The expression of Fgf4 in embryos at an early blastula stage, the128-cell stage, was examined by whole mount in situ hybridization (a). The expression of Fgf4 and lrdr1, a marker gene for Kupffer′s vesicle, in embryos at an early stage of segmentation (12 hpf) was also examined by whole mount in situ hybridization. Fgf4 was predominantly expressed in the posterior notochord and Kupffer′s vesicle and its peripheral region. Arrows indicate Kupffer′s vesicle (b?e). Fgf4 was also expressed in the MHB, pharyngeal arch and tail bud at 24 hpf (f) and in the MHB, fin buds and somites at 48 hpf (g). Lateral views (b, d, f, g); dorsal views (c, e). Scale bars = 200 μm. |
|
The formation of Kupffer′s vesicle in Fgf4 knockdown embryos. Kupffer′s vesicle in Fgf4 MO1-injected embryos at 12 hpf was morphologically normal (100%, n = 32) (a, b). The expression of lrdr1, which is essential for cilia to function in Kupffer′s vesicle, was examined by whole mount in situ hybridization. The expression was also unaffected in Fgf4 MO1-injected embryos at 10 hpf (93.3%, n = 30) (c, d). (a?d) Dorsal views of the tail bud. A scale bar = 200 μm. Cilia exist on the cells in Kupffer′s vesicle. The expression of acetylated tubulin, a marker protein for cilia, was examined by immunohistochemistry using an anti-acetylated tubulin antibody. The size of Kupffer′s vesicle and the number of cilia of Kupffer′s vesicle in Fgf4 MO-injected embryos were not significantly affected (100%, n = 30); however, the average length of cilia of Kupffer′s vesicle in Fgf4 MO-injected embryos (3.27 ± 1.06 μm, n = 204) was significantly shorter than that in wild-type embryos (4.41. ± 0.91 μm, n = 214) (e, f). These results indicate that Fgf4 is not essential for the morphology and function of Kupffer′s vesicle. A scale bar = 20 μm. EXPRESSION / LABELING:
PHENOTYPE:
|
|
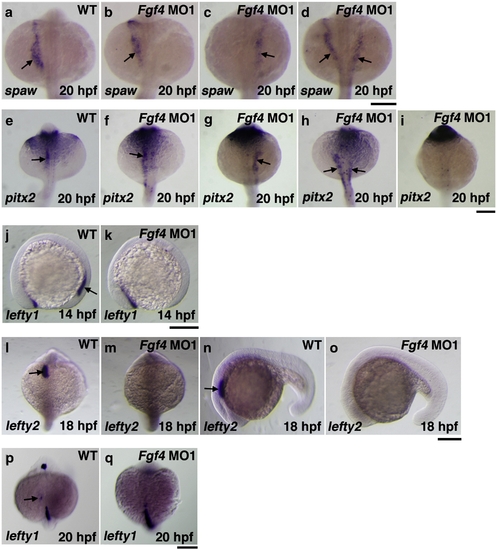
Asymmetric gene expression in Fgf4 knockdown embryos. The expression of southpaw in embryos at 20 hpf was examined by whole mount in situ hybridization. The expression was expanded anterior-laterally in the left LPM in wild-type embryos (a). In contrast, the expression of southpaw in Fgf4 MO1-injected embryos at 20 hpf was randomized (normal (33.3%, n = 30); reverse (23.3%, n = 30); bilateral (43.3%, n = 30)) (b?d). In wild-type embryos, the expression of pitx2 was detected in the LPM at 20 hpf (e). In contrast, the expression of pitx2 was randomized (normal (36.7%, n = 30); reverse (23.3%, n = 30), bilateral (20%, n = 30); absence (20%, n = 30)) (f?i). Although the expression of lefty1 was detected in the posterior notochord of wild-type embryos at 14 hpf, it was not detected in Fgf4 MO1-injected embryos (81.2%, n = 32) (j, k). The expression of lefty2 and lefty1 in the left cardiac field was examined in embryos at 18 hpf and 20 hpf, respectively. The expression of lefty2 was strongly detected in the cardiac field of wild-type embryos (l, n). However, the expression was not detected in Fgf4 MO1-injected embryos (93.3%, n = 30) (m, o). The expression of lefty1 was weakly detected in the cardiac field of wild-type embryos (p). The expression was not detected in Fgf4 MO1-injected embryos (86.7%, n = 30) (q). Arrows indicate positive staining. Scale bars = 200 μm. EXPRESSION / LABELING:
PHENOTYPE:
|
|
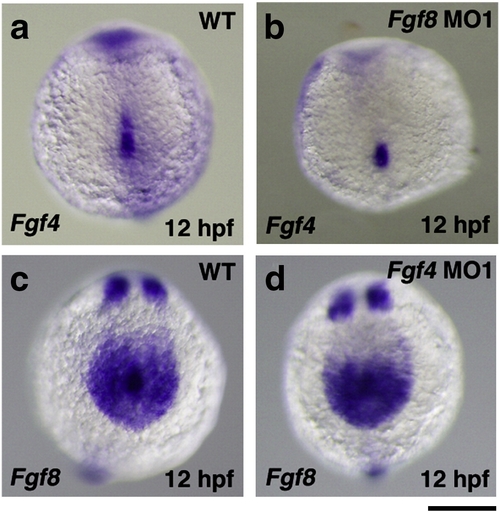
The expression of Fgf4 and Fgf8 in Fgf8 and Fgf4 knockdown embryos, respectively. The expression of Fgf4 in Kupffer′s vesicle and its peripheral region in wild-type and Fgf8 MO-injected embryos was examined by whole mount in situ hybridization. The expression of Fgf4 was detected in Fgf8 MO-injected embryos (81.3%, n = 32) (a, b). The expression of Fgf8 in Kupffer′s vesicle and its peripheral region was also examined. The expression of Fgf8 was also essentially unchanged in Fgf4 MO1-injected embryos (100%, n = 32) (c, d). A scale bar = 200 μm. EXPRESSION / LABELING:
|
Reprinted from Developmental Biology, 332(1), Yamauchi, H., Miyakawa, N., Miyake, A., and Itoh, N., Fgf4 is required for left-right patterning of visceral organs in zebrafish, 177-185, Copyright (2009) with permission from Elsevier. Full text @ Dev. Biol.