Fig. 1
|
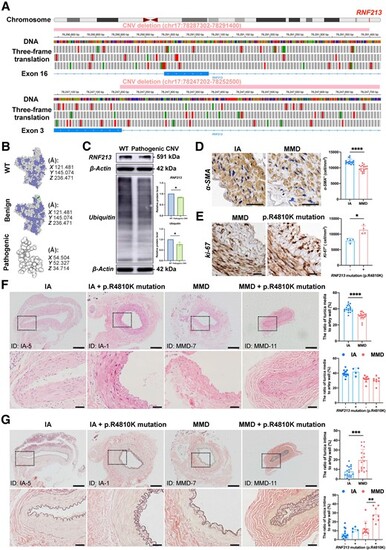
RNF213 loss-of-function mutations exist in some MMD patients and are positively associated with intimal hyperplasia in MMD patients. (A) The integrative genomics viewer (IGV) application showed a benign copy number variant (CNV) deletion (chr17:78287302-78291400) that completely damages the genomic sequence of exon 16 and a likely pathogenic CNV deletion (chr17:78247202-78252500) that partly damages the genomic sequence of exon 3 in RNF213. The green boxes indicate a start codon, while the red boxes indicate termination codons in the three-frame translation. (B) The protein structure of the wild-type (WT) RNF213 protein (Q63HN8) was modelled with 100.0% confidence based on the single highest-scoring template of the well-known mouse Rnf213 (c7oikA). The benign CNV deletion of RNF213 showed a high identity (99.42%) with WT, while the clinically relevant CNV deletion of RNF213 showed a very low similarity (30.48%) with WT, and only 69 residues were modelled with 43.4% confidence using SWISS-Model and Phyre2. (C) Quantitative analysis of RNF213 and ubiquitin protein levels in one intracranial aneurysm (IA) patient and moyamoya disease (MMD) patient carrying the pathogenic RNF213 mutation of exon 3 (RNF213, n = 4, 1.00 ▒ 0.00 versus 0.84 ▒ 0.03, P = 0.0286; ubiquitin, n = 4, 1.00 ▒ 0.00 versus 0.78 ▒ 0.09, P = 0.0286). (D) The vascular smooth muscles were then stained with an ?-SMA antibody, and the region of interest (ROI)-based quantitative analysis of ?-SMA+ cells in the tunica media indicated a relative loss of smooth muscle in MMD patients (n = 18, 11944.81 ▒ 783.90 versus 9796.53 ▒ 962.64 cell/mm2, P < 0.0001). Scale bar = 20 Ám. (E) The density of Ki-67 was quantitatively analysed in the tunica intima, which indicated the presence of increased number of Ki-67+ cells in MMD patients with RNF213 mutation (n = 4, 7944.90 ▒ 706.61 versus 11 373.69 ▒ 1348.96 cell/mm2, P = 0.0286). Scale bar = 20 Ám. (F) Haematoxylin and eosin staining was used for morphometric analysis of the vascular wall structure of donor superficial temporal artery (STA) sections from MMD and IA patients. MMD patients showed a significantly thinner tunica media than IA patients (n = 18, 31.09 ▒ 4.35 versus 40.22 ▒ 4.33%, P < 0.0001). No significant difference in tunica media thickness was observed between MMD patients with RNF213 mutation and those without RNF213 mutation (n = 8 versus 10, 30.19 ▒ 4.86 versus 31.82 ▒ 4.01%, P = 0.3599). A significant difference in tunica media thickness was not observed between IA patients carrying RNF213 mutation and those without RNF213 mutation (n = 4 versus 14, 42.00 ▒ 5.27 versus 39.71 ▒ 4.11%, P = 0.4418). Scale bar = 200 Ám (top); scale bar = 40 Ám (bottom). (G) Weigert?s resorcin-fuchsin staining was applied for morphometric analysis of the internal elastic membranes in MMD and IA patients. MMD patients showed significantly thicker internal elastic membranes than IA patients (n = 18, 19.53 ▒ 11.16 versus 7.68 ▒ 5.12%, P = 0.0007). MMD patients carrying RNF213 mutation presented with a significantly greater tunica intima thickness than MMD patients without RNF213 mutation (n = 8 versus 10, 28.29 ▒ 7.87 versus 12.53 ▒ 8.02%, P = 0.0014). No significant difference in tunica intima thickness was observed between IA patients with RNF213 mutation and those without RNF213 mutation (n = 4 versus 14, 11.55 ▒ 2.54 versus 6.58 ▒ 5.18%, P = 0.0791). Scale bar = 200 Ám (top); scale bar = 40 Ám (bottom). Each dot represents one sample. The means ▒ SDs are shown. The dashed line indicates the ROI. *P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001. |