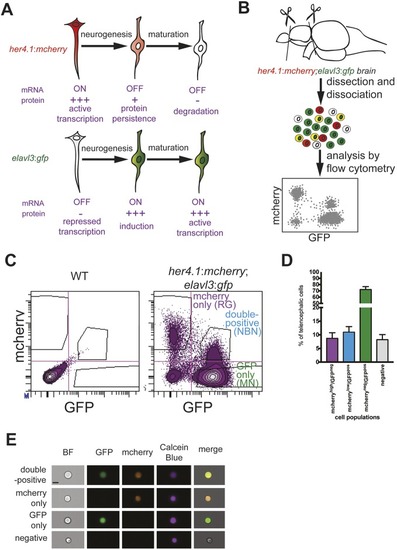
A double-reporter line for identification and quantification of radial glia-derived newborn neurons in the adult zebrafish brain. (A) Model for the lineage-trace paradigm. NBNs stop to transcribe her4.1-driven mcherry, but inherit the protein from their RG precursors (top). At the same time, NBNs induce transcription of the neuronal marker elavl3:gfp (bottom), allowing the identification of NBNs as mCherry/GFP-double-positive cells in double reporter fish. (B) Scheme of the analytical workflow. Forebrains are removed from double reporter fish and dissociated, and cell types are analyzed by flow cytometry. (C) Representative FACS plots showing forebrain cells from wild-type (left) and her4.1:mcherry;elavl3:gfp double reporter fish (right). There are clearly separated populations of mCherryhigh/GFPneg RG and mCherrylow/GFPpos NBNs. (D) Quantification of cells in the four quadrants shown in C, revealing that NBNs can be found in a frequency comparable with RG and represent circa 10% of forebrain cells. n=5. Data are mean±s.e.m. (E) Representative single cells imaged in flow cytometry for mCherrypos/GFPpos (first row), mCherrypos/GFPneg (second row), mCherryneg/GFPpos (third row) and mCherryneg/GFPneg (fourth row). At least 20 cells were analyzed and no doublets were seen, excluding the double-positive cells that represent doublets. Scale bar: 7 µm.
|