- Title
-
A zebrafish model of growth hormone insensitivity syndrome with immune dysregulation 1 (GHISID1)
- Authors
- Heidary, S., Awasthi, N., Page, N., Allnutt, T., Lewis, R.S., Liongue, C., Ward, A.C.
- Source
- Full text @ Cell. Mol. Life Sci.
|
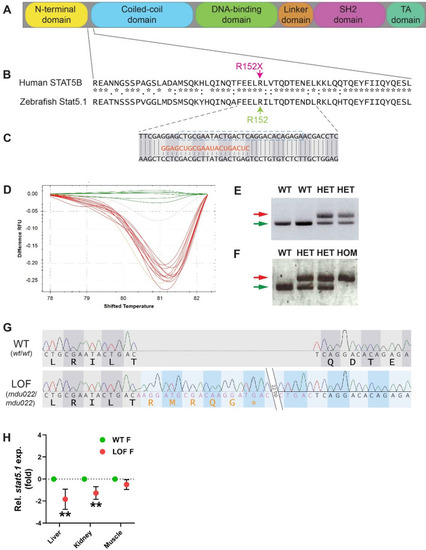
Generation of a zebrafish Stat5.1 mutation mimicking those in human STAT5B associated with GHISID1. A Schematic diagram of the STAT5B protein domain architecture consisting of N-terminal (yellow), coiled-coil (blue), DNA-binding (green), linker (orange), SH2 (pink), and transactivation (TA) (dark green) domains. B Alignment of human STAT5B with zebrafish stat5.1 sequences around the site of the R152X mutation (green) seen in human GHISID1, showing identical (*), highly similar (:) and similar (.) amino acids. C Nucleotide sequence of zebrafish stat5.1 genomic DNA targeted using CRISPR/Cas9 with the gRNA (orange) shown and the coding phase indicated. D HRM analysis of a sample of embryos injected with stat5.1 gRNA and Cas9, with wild-type embryos in green and other colours representing potential mutants. |
|
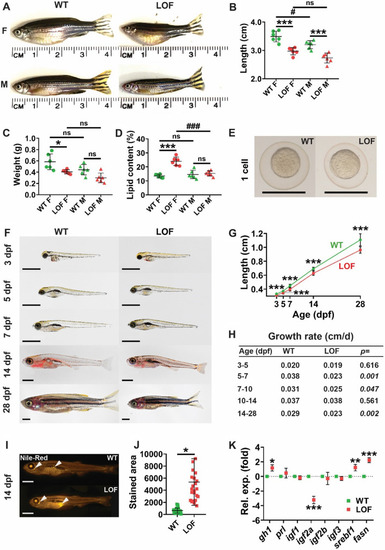
Effect of LOF Stat5.1 mutation on growth and adiposity. A–D Assessment of adult size and body composition in female (F) and male (M) homozygous stat5.1wt/wt wild-type (WT) and stat5.1mdu022/mdu022 loss-of-function (LOF) mutants at 5 months post-fertilisation (mpf), presenting representative images (A) along with quantification of standard length (B), wet weight (C) and lipid content (D), showing values for individual fish as well as mean ± standard deviation (SD) with statistical significance indicated (n = 6). E–H Growth trajectory during early life, presenting light microscopic images of representative WT and LOF individuals at the 1 cell stage (E) and at the indicated times post-fertilisation (F), along with quantification of length showing mean ± SD and statistical significance (n = 10–33) (G) and relative growth rate in the indicated age ranges calculated from these data with p-values shown (H). Scale bar = 1 mm. I–J Lipid analysis in juveniles, presenting fluorescent images of representative 14 dpf WT and LOF stained with Nile Red (I) and quantification of the area of Nile Red staining showing individual values as well as mean ± SD, with statistical significance indicated (n = 24) (J). Scale bar = 1 mm. K Expression analysis of growth and metabolic genes in 28 dpf juveniles (K) using qRT2-PCR on WT and LOF samples. Data for the indicated genes were normalised to actb and represented as relative fold-change compared to WT, with mean ± SD shown and statistical significance indicated (n = 6) EXPRESSION / LABELING:
|
|
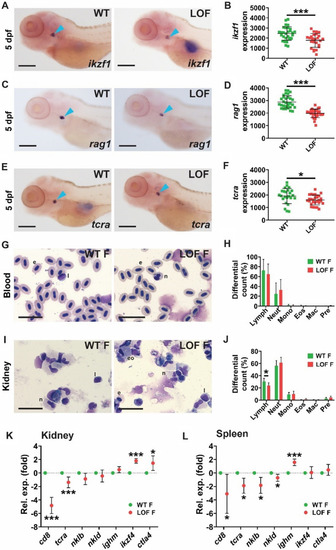
Effect of LOF Stat5.1 mutation on lymphoid cells. A–F Assessment of early lymphopoiesis in 5 dpf homozygous stat5.1wt/wt wild-type (WT) and stat5.1mdu022/mdu022 loss-of-function (LOF) mutant embryos using WISH analysis, presenting representative individuals for ikzf1(A), rag1(C) and tcra(E), along with quantification of the relative area of expression for ikzf1(B), rag1(D) and tcra(F), showing results for individual embryos along with mean ± SD and statistical significance (n = 25–31). Scale bar = 200 μm. G–J. Analysis of blood and immune cells in adults, presenting images of representative Giemsa-stained blood (G) and kidney marrow (I) smears from 5 month post-fertilisation WT and LOF female fish and quantification of indicated cell populations in blood (H) and kidney marrow (J) showing mean and SD with statistical significance indicated (n = 6). Scale bar = 20 μm. K–L Expression analysis of immune genes in adults using qRT2-PCR on kidney marrow |