- Title
-
Delta/Jagged-mediated Notch signaling induces the differentiation of agr2-positive epidermal mucous cells in zebrafish embryos
- Authors
- Lu, Y.F., Liu, D.W., Li, I.C., Lin, J., Wang, C.M., Chu, K.C., Kuo, H.H., Lin, C.Y., Yih, L.H., Jiang, Y.J., Hwang, S.L.
- Source
- Full text @ PLoS Genet.
|
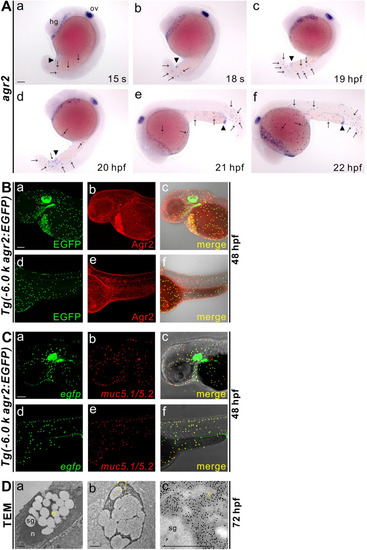
Characterization of agr2+ epidermal mucous cells. (A). Developmental expression pattern of agr2 in the epidermis from 15 s to 22 hpf. hg, hatching gland; ov, otic vesicle. Arrows, EMCs. Arrowhead, anus. Scale bar, 100 ?m. (B). Colocalization of Agr2 (red) and EGFP (green) was detected in the trunk and yolk regions of Tg(-6.0 k agr2:EGFP) embryos at 48 hpf. Scale bar, 100 ?m. (C). egfp (green) is coexpressed with muc5.1/muc5.2 (red) in Tg(-6.0 k agr2:EGFP) embryos at 48 hpf. Scale bar, 100 ?m. (D). Ultrastructure of EMCs and GFP immuno labeling in Tg(-6.0 k agr2:EGFP) embryos at 72 hpf. n, nucleus; sg, secretory granules. Yellow arrowhead, endoplasmic reticulum. Yellow square box indicates enlarged area. Scale bars, 1 ?m. Scale bar for enlarged image (c), 0.5 ?m. EXPRESSION / LABELING:
|
|
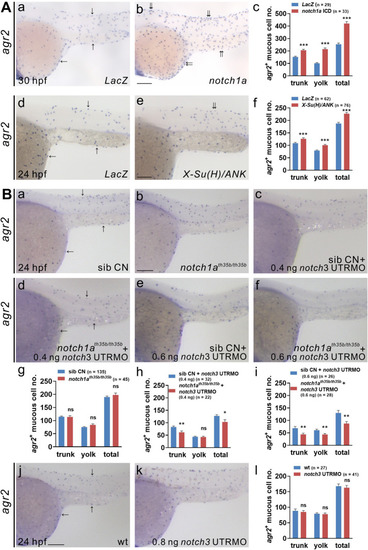
Notch signaling is required for the differentiation of agr2+ epidermal mucous cells.(A).Significant reductions in agr2+ EMC numbers were detected in two mib homozygous mutants compared to sibling wild-type embryos at 24 hpf. (B). Larger reductions in agr2+ EMC numbers were detected in embryos treated with ?-secretase inhibitor (MK0752) from 10?15 hpf, 10?16 hpf and 10?17 hpf than in those treated from 12?17 hpf. Arrows, EMCs. Scale bars, 100 ?m. Mean ± SEM. Student?s t-test. *p<0.05; **p<0.01; ***p<0.001; ns, not significant. Underlying data are available in S1 Data. |
|
Notch1a and Notch3 receptors participate in the differentiation of agr2+ epidermal mucous cells. (A). notch1a or X-Su(H)-ANK-overexpressing embryos display significant increases in agr2+ EMC numbers in the trunks and yolks compared to LacZ-overexpressing embryos at 30 or 24 hpf. (B). Similar agr2+ EMC numbers in the trunks and yolks were detected in notch1a homozygous mutants or notch3 morphants compared to sibling wild-type or wild-type embryos at 24 hpf. Substantial reductions in agr2+ EMC numbers in the trunks and yolks are detected in notch1a homozygous mutants injected with 0.4 or 0.6 ng notch3 UTRMO compared to sibling wild types injected with the same amount of notch3 UTRMO at 24 hpf. Arrows, EMCs. Scale bars, 100 ?m. Mean ± SEM. Student?s t-test. *p<0.05; **p<0.01; ***p<0.001; ns, not significant. Underlying data are available in S1 Data. |
|
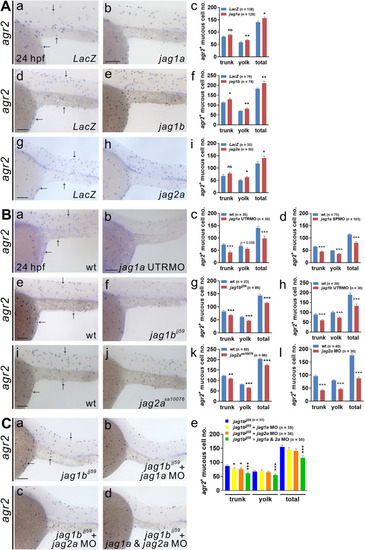
Jagged1a, Jagged1b and Jagged2a ligands participate in the differentiation of agr2+ epidermal mucous cells.(A).Significant increases in agr2+ EMC numbers in the trunks and yolks were detected in embryos overexpressing jag1a, jag1b or jag2a mRNA compared to LacZ mRNA-overexpressing embryos at 24 hpf. (B). Significant reductions in agr2+ EMC numbers in the trunks and yolks were detected in embryos injected with jag1a UTRMO, jag1a SPMO, jag1b UTRMO or jag2a MO, as well as jag1b and jag2a homozygous mutants compared to wild-type embryos at 24 hpf. (C). Further reductions in agr2+ EMC numbers in the trunks and yolks were observed in jag1b homozygous mutant embryos injected with jag1a and jag2a MOs compared to those injected with jag1a or jag2a MO alone or un-injected jag1b homozygous mutant embryos at 24 hpf. Arrows, EMCs. Scale bars, 100 ?m. Mean ± SEM. Student?s t-test. *p<0.05; **p<0.01; ***p<0.001; ns, not significant. Underlying data are available in S1 Data. |
|
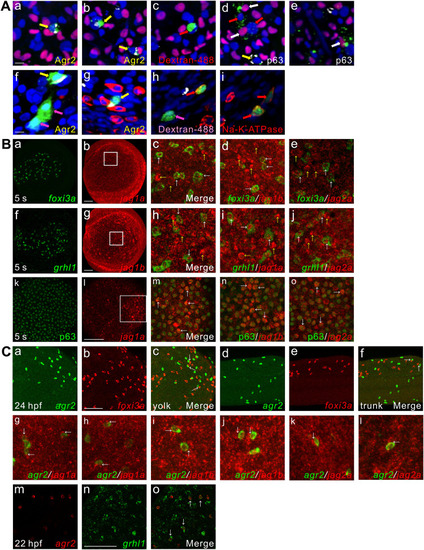
agr2+ EMCs and keratinocytes or ionocytes are derived from common ectodermal precursors, coexpression of jag1a/jag1b/jag2a with foxi3a, grhl1 or p63, agr2 is expressed next to foxi3a or jag1a/ jag1b/jag2a and coexpression of agr2 and grhl1. (A). agr2+ EMCs and keratinocytes or ionocytes are derived from common ectodermal precursors. Yellow arrows (a, b, d, f, g) indicate Agr2+ and dextran+ (dextran-488) cells. White arrows (d, e) indicate P63+ and dextran+ cells. Red arrows (c, d) and pink arrows (f, h) indicate only dextran+ cells. Red arrows (h, i) indicate Na+-K+-ATPase+ and dextran+ cells. Scale bars, 10 ?m. (B). Coexpression of jag1a/jag1b/jag2a with foxi3a, grhl1 or p63 detected at 5 s. White square boxes indicate enlarged area. White arrows indicate examples of colocalized cells. Yellow arrows indicate example of jag1a-, jag1b- or jag2a-expressing cells. Images (k-o) were derived from 3 to 4 Z-stacks. (C). Some foxi3a+ cells locate next to agr2+ EMCs in the trunks and yolks of embryos at 24 hpf. Some jag1a-, jag1b- or jag2a-expressing cells were located next to agr2+ EMCs in the epidermis of embryos at 22 hpf. Coexpression of grhl1 and agr2 was observed in embryos at 22 hpf. White arrows (c, f, g-l) indicate examples of agr2+ EMCs located adjacent to foxi3a+ or jag1a-, jag1b- or jag2a-expressing cells. White arrows (o) indicate examples of colocalized cells. Scale bars, 100 ?m. Underlying data are available in S1 Data. |
|
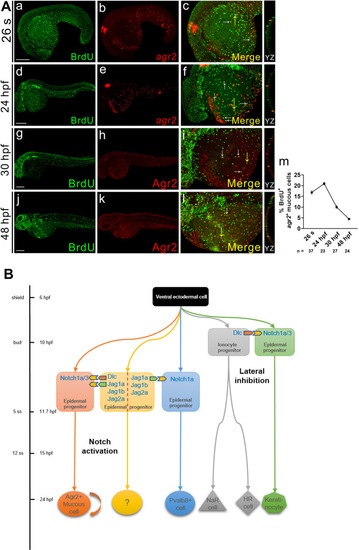
agr2+ EMC population is maintained by proliferation.(A). Double immunofluorescence and in situ hybridization or double immunofluorescence reveals colocalization of BrdU (green) and agr2/Agr2 (red) in the yolks of embryos at 26 s, 24, 30 and 48 hpf. BrdU labeling assay revealed that agr2+ EMCs proliferate after differentiation. The highest proliferation rate was detected at 24 hpf. White arrows indicate BrdU+agr2+ EMCs. Yellow arrows indicate positions of YZ projections of confocal images. Mean ± SEM. Scale bars, 100 ?m. (B). A model depicts the timing at which sequential specification of ionocytes/keratinocytes and agr2+ EMCs/pvalb8-positive cells occurring in zebrafish embryos. The previously-explored specification of ionocytes and keratinocytes via Dlc-mediated Notch1a/3 lateral inhibition occurs at late gastrulation stage, while the newly-identified induction of agr2+ EMCs by Dlc and Jag1a/1b/2a-mediated activation of Notch1a/3 signaling and the induction of pvalb8-positive cells via Jag1a/Jag1b/Jag2a-mediated activation of Notch1a signaling occur during segmentation stage. Underlying data are available in S1 Data. EXPRESSION / LABELING:
|