- Title
-
Recurrent NFIA K125E substitution represents a loss-of-function allele: Sensitive in vitro and in vivo assays for nontruncating alleles
- Authors
- Uehara, T., Sanuki, R., Ogura, Y., Yokoyama, A., Yoshida, T., Futagawa, H., Yoshihashi, H., Yamada, M., Suzuki, H., Takenouchi, T., Matsubara, K., Hirata, H., Kosaki, K., Takano-Shimizu, T.
- Source
- Full text @ Am. J. Med. Genet. A
|
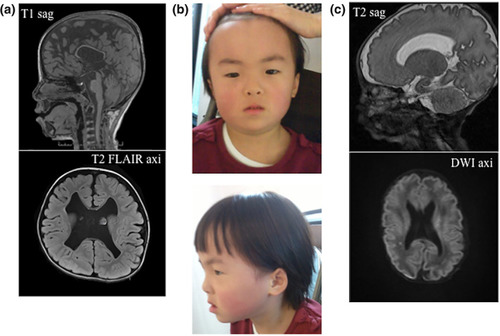
Clinical characteristics of two patients with the same |
|
Amino acid sequence alignment of the DNA-binding domains of human NFIA with its orthologs and paralogs. Sequences and domain definitions were obtained from UniProt (The UniProt Consortium, 2019). HspA: human NFIA (Q12857, residues 1?194); HspB: human NFIB (O00712, 1?195); HspC: human NFIC (P08651, 1?195); HspX: human NFIX (Q14938, 1?194); Mmu: mouse Nfia (Q02780, 24?217); Xtr: frog nfia (A0A6I8RQ42, 1?194); Dre: zebrafish nfia (F1R2R8, 1?193); Dme: fly NfI (Q86P06, 19?212); and Cel: nematode nfi-1 (Q09631, 44?237). The K125E mutation site is indicated by an arrow; 6th and 7th mutagenized sites in Armentero et al. (1994) are underlined |
|
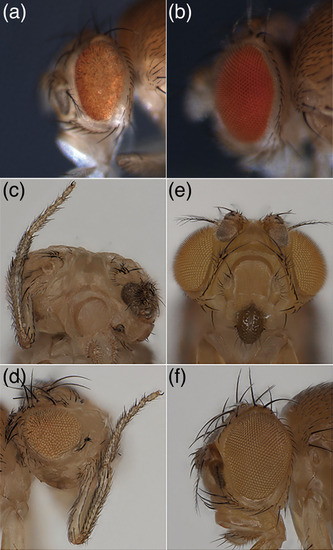
Phenotypes of ectopic expression of |
|
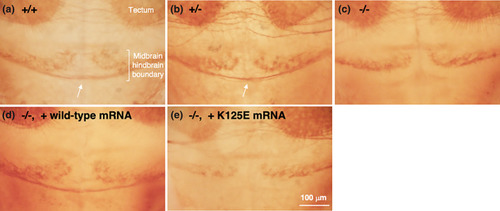
Commissural defects in PHENOTYPE:
|
|
|