- Title
-
Zebrafish Wtx is a negative regulator of Wnt signaling but is dispensable for embryonic development and organ homeostasis
- Authors
- Große, A., Perner, B., Naumann, U., Englert, C.
- Source
- Full text @ Dev. Dyn.

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. |
|
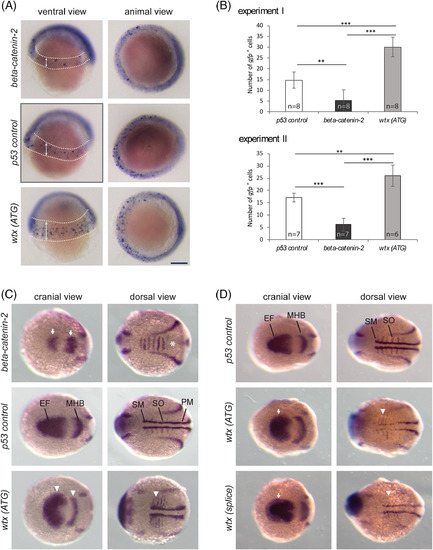
Wtx inhibits the canonical Wnt pathway in early zebrafish development. Whole mount in situ hybridization to detect mRNA expression changes indicative of modulated Wnt/??catenin signaling. The wtx depletion phenotypes were compared with p53?control MO?injected embryos and beta?catenin?2 MO?injected embryos. A, Examination of gfp expression in zebrafish embryos heterozygous for a ??catenin responsive transgene Tg(TOP:dGFP) at shield stage (6 hpf). Embryos are shown from a ventral view with the animal pole at top and from an animal view with dorsal to the right. Depletion of ??catenin2 resulted in reduced transgene expression whereas embryos injected with the wtx MO displayed increased number of ??catenin responsive cells in an expanded area within the ventrolateral margin. GFP?positive cells are recognizable as blue spots inside the areas which are marked with dashed lines in the ventral views; arrows illustrate the expansion of the transgene expressing area in wtx morphants. The diffuse staining around the edge of the embryo is not specific. B, Quantification of the data shown in (A). Images were blinded and the numbers of gfp positive cells were counted. Results from two independent experiments are shown. Error bars represent SD. P values according to the two?tailed t?test after Bonferroni correction: ** P?<?.01, *** P?<?.001. C and D, Expression analysis of markers for lateral mesoderm and neuroectoderm by using a pax2a/myod1/six3b probe mixture on 12 hpf zebrafish embryos. Embryos are shown in a cranial and dorsal view with anterior to the left. C, Knockdown of ??catenin2 caused a lateral shortening of the pax2a expression domain in the MHB and reduced expression of six3b in the optic primordia (white arrows). The myod1 domain in the somites is fused (white asterisk) and absent in the somitic and presomitic mesoderm. In contrast wtx morphants show expanded expression of pax2a, myod1 and six3b(white arrowheads). For ?beta?catenin?2, ?p53 control? and ?wtx (ATG)? 6, 3 and 11 embryos were analyzed, respectively. (D) Wtx (ATG) and wtx (splice) morphants show expanded expression of six3b (white arrows) and myod1 (white arrowheads). For ?p53 control?, ?wtx (ATG)? and ?wtx (splice)? 7, 13 and 9 embryos were analyzed, respectively. EF: eye field; MHB: midbrain?hindbrain boundary; PM: presomitic mesoderm; SM: somitic mesoderm; SO: somites. Scale bar represents 150??m EXPRESSION / LABELING:
PHENOTYPE:
|
|
Wtx inactivation is not associated with developmental anomalies. A, Structure of the wtx gene locus on zebrafish chromosome 5. The coding sequence is indicated as a black box, grey boxes depict untranslated sequences. Binding sites of both TALEN pairs are indicated by arrowheads. Arrows (F, R) show primers used for amplification. B, Sequences of selected wtx mutations in F1 fish derived from TALEN?1 injections close to the transcriptional start site and from TALEN?1/TALEN?2 co?injection?induced segmental deletions. Deletions are indicated by dashes. Underlined sequences correspond to the left arm and the right arm of TALENs. The total number of deleted nucleotides in mutant alleles is given in brackets. C, Predicted consequences of TALEN?induced mutations for protein sequence. The mutations wtx del1 and wtx del2 are predicted to cause reading frame shifts resulting in premature translation termination codons at amino acid position 20 and 16, respectively. The mutation wtx del3 is predicted to delete 421 amino acids and therefore five of six conserved sequence blocks (orange, yellow, red, green, blue, purple) and thus nearly half of the protein. MBD, membrane binding domain; aa, amino acids. D, Localization of wild type and N?terminally truncated Wtx. ZF4 cells were transiently transfected with the respective expression vector. HA?epitope (green) was detected 48?hours post transfection (hpt). DAPI (blue) was used for nuclear staining. White arrow indicates membrane localization of wtx?HA. Scale bars represent 10 ?m. (right) Western blot against HA?epitope of ZF4 cells transfected with the respective expression vector (48 hpt). E, Representative brightfield images of wild type and homozygous wtx del3 mutant zebrafish 48?hpf and F, 7 dpf. Mutant wtx alleles do not result in overt embryonic phenotypes. D, Wild type and homozygous wtx del3 mutant embryos, bearing Tg(wt1b:EGFP) transgene. Transgene expression (green) demarks the pronephros, consisting of glomeruli (G) and associated tubuli (T). E, At 7 dpf calcein staining (green) indicates craniofacial bone structures of wild type and wtx del3 mutant zebrafish larvae (head region). For orientation, E depicts eye, O demarks opercle |
|
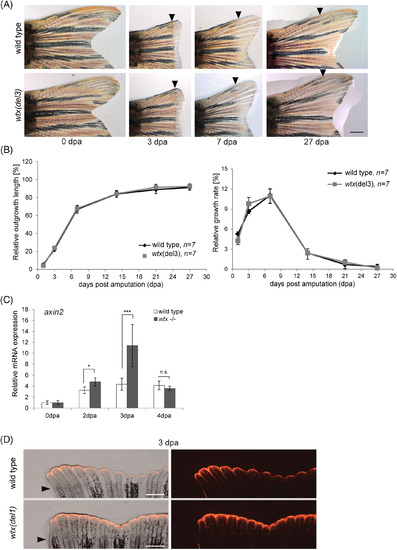
Loss of wtx increases Wnt/??catenin activity during caudal fin regeneration. A, Images of representative regenerating tail fins (dorsal side) in wild type and in wtx del3 mutant zebrafish after 50% amputation. Re?growing fins were monitored for 27?days. Displayed are 0, 3, 7 and 27 dpa. Black arrowheads indicate the amputation plane. Scale bar represents 1 mm. B, Relative outgrowth kinetics of length and growth rate, measured over 27?days of regeneration. Displayed are mean values. Error bars represent SD. C, Relative mRNA expression of axin2 in uninjured fins (0 dpa) and regenerating fin tissues at 2 dpa, 3 dpa and 4 dpa of wild type and homozygous wtx mutants ( wtx del1 , wtx del2 and wtx del3 ) fish using qRT?PCR. Gene expression was normalized to uninjured fin (0 dpa) and to the reference gene ef1. Three samples per time point were analyzed consisting of 3 to 4 animals each. Error bars display SE of the mean (SEM). P?values according to the two?tailed Student's t?test: * P?<?.05; *** P?<?.001; n.s.: not significant. D, Fin regenerates of wtx(del1) mutant fish and wild type controls at 3 dpa following 50% fin amputation in a Tg(TCFsiam:mCherry) background shows expanded fluorescent signals (red) in wtx mutants (exposure time: 3 seconds). Six wild type and six mutant animals were analyzed. Black arrowheads indicate amputation plane. Scale bars represent 500??m |
|
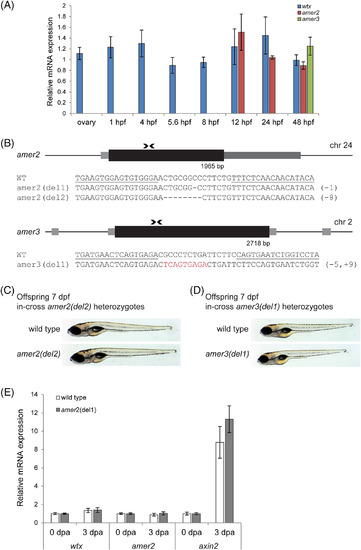
Mutant amer2 and amer3 alleles are not associated with embryonic anomalies. A, Relative mRNA levels of wtx, amer2 and amer3 in female ovaries and in zebrafish embryos of different developmental stages (1 hpf ? 48?hpf) of wild type and homozygous wtx mutants ( wtx del1 , wtx del3 ), analyzed by qRT?PCR. Gene expression was normalized to ef1a. Wild type was set as 1; the ratio between mutant/wild type is shown. N = 3 for wild type and wtx del3 ovaries, as well as for pooled embryos (expression data of wtx del1 and wtx del3 were combined). The error bars indicate SEM. B, Structure and partial sequence of the amer2 gene locus on zebrafish chromosome 24 and the amer3 gene locus on zebrafish chromosome 2. The coding sequences are indicated as black boxes, grey boxes depict untranslated sequences. Binding sites of the TALENs are indicated by arrowheads. Below the schemes sequences of selected amer2 and amer3mutations in F1 fish are shown. Deletions are indicated by dashes. Red letters indicate nucleotide insertions. Underlined sequences correspond to the left and the right arm of the TALENs. In brackets the total number of deleted or inserted nucleotides in mutant alleles are indicated. C and D, Representative brightfield images of 7 dpf homozygous mutant zebrafish larvae resulting from an in?cross of heterozygous fish with TALEN induced B, amer2 del2 and C, amer3 del1 mutation. E, Relative mRNA expression of wtx, amer2 and axin2 in uninjured fin tissue at 3 dpa measured by qRT?PCR. Gene expression was normalized to uninjured fin (0 dpa) and to the reference gene ef1a. Tissues of three animals were pooled. Error bars represent SEM |
|
Wtx/amer2/amer3 triple mutant embryos display dorsalization similar to wtxmorphants. Whole mount in situ hybridization for expression analysis of markers for lateral mesoderm and neuroectoderm. A mixture of pax2a, myod1 and six3bprobes was applied to wild type and wtx/amer2/amer3 triple mutant embryos. The mutants display an expansion of the respective expression domains (white arrowheads) in the midbrain?hindbrain boundary ( pax2a), optic primordia ( six3b) and in the somites ( myod1). Triple mutants injected with a wtx morpholino (ATG) have similarly expanded expression domains. Embryos were analyzed at 12 hpf and are shown with anterior to the left in a cranial as well as in a dorsal view. For ?wild type uninjected?, ?triple mutant uninjected? and ?triple mutant wtx (ATG) MO? 17, 19, and 13 embryos were analyzed, respectively. EF, eye field; MHB, midbrain?hindbrain boundary; PM, presomitic mesoderm; SM, somatic mesoderm; SO, somites. Scale bar represents 200??m EXPRESSION / LABELING:
PHENOTYPE:
|