- Title
-
Genetic Dissection of Dual Roles for the Transcription Factor six7 in Photoreceptor Development and Patterning in Zebrafish
- Authors
- Sotolongo-Lopez, M., Alvarez-Delfin, K., Saade, C.J., Vera, D.L., Fadool, J.M.
- Source
- Full text @ PLoS Genet.
|
ljrp23ahub mutants display an increased number and uniform distribution of rod photoreceptors. (A) Confocal immunofluorescent images labeled for rods (red) and UV-sensitive cones (green) from WT, ljrp23ahub, lorp25bbtl and homozygous lorp25bbtl/ljrp23ahub retinas at 4 days-post-fertilization (dpf). WT larvae show asymmetric rod distribution in central and dorsal retina and a uniform distribution of UV-sensitive cones across the entire retina. ljrp23ahub and lorp25bbtl mutants demonstrate an increased number and uniform arrangement of rods but fewer UV-sensitive cones labeled in lorp25bbtl. Double-mutant retinas display higher rod labeling and few UV-sensitive cones. (B) Graph showing the average number of rods per unit area dorsal to the optic nerve (WT, n = 5; ljrp23ahub, n = 5; lorp25bbtl, n = 5; ljrp23ahub/lorp25bbtl, n = 5). Significantly-different means by one-way ANOVA with Tukey?s post-hoc test, b vs c, p<0.05, all other comparisons p<0.0001. (C) Graph showing the average number of UV-sensitive cones per unit area from samples used in 1B, a vs b, p<0.0001. (D) Graph showing the average number of rods per unit area (WT, n = 5; ljrp23ahub/+, n = 6; ljrp23ahub, n = 5); a vs b, p<0.05, all other comparisons p<0.0001. (E) Flat mount views of confocal immunofluorescent images labeled for red-sensitive (brighter signal) and green -sensitive (dimmer signal) cones from WT and ljrp23ahub retinas at 4 dpf. ljrp23ahub mutants maintain the alternating arrangement of red- and green-sensitive cones. (F) Graph showing the average number of red- and green-sensitive cones per unit area (WT, n = 6; ljrp23ahub, n = 6). No significant differences are observed, Student?s t test, p>0.05. Error bars represent SD. |
|
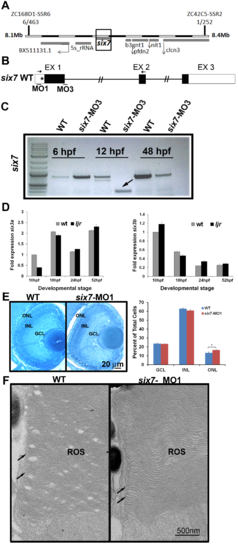
six7 knockdown phenocopies the increase and uniform distribution of rods from ljrp23ahub mutants. (A) Frequency of SNPs across chromosome 7 relative to the TL reference genome (danRer7) calculated from whole-genome sequencing of a pool of DNA extracted from 118 ljrp23ahub mutants where TL is the reference background. Below, 75 kb view of the SNP-depleted region on chromosome 7. A 2.4 kb region depleted of uniquely-aligning reads is highlighted in teal. Shown are tracks for H3K4me1 [63], read alignments from whole-genome sequencing of ljrp23ahub mutants, and multiz-based sequence conservation UCSC genome browser tracks of six7 across four fish species and Six3 in frog, human, and mouse [72]. Gel electrophoresis of PCR products of six7 exon 1 and the upstream genomic region amplified from AB, TL and ljrp23ahub genomic DNA. (B) Whole mount in situ hybridization shows six7 expression confined to retinal neuroblasts and differentiating ONL spatially and temporally with photoreceptor genesis. Dorsal is up. (C) Quantitative RT-PCR (qRT-PCR) performed on mRNA from control (WT) and ljrp23ahub embryos at 10 hpf, 18 hpf, 24 hpf and 52 hpf reveal down regulation of six7 expression in ljrp23ahub at 52 hpf. Relative transcript abundance was normalized to actin levels and is presented as the mean fold change in expression relative to 10 hpf controls (n = 30 embryos per group). All the real-time PCR experiments were carried out in triplicate. Significant differences observed at 18 hpf and 52 hpf, Student?s t test, *p<0.05. (D) Retinal cryosection from 4 dpf un-injected control WT and six7-MO1 injected embryos immunolabeled for rods (4C12, green). six7-morphants display an increase in the number of rods as detected in ljrp23ahub. Note the lack of gaps in rod distribution in the central retina of six7-knockdown larvae. (E) Graph showing dosage dependent increase in the average number of rods per unit area dorsal to the optic nerve of WT and six7-MO1 injected embryos (WT un-injected, n = 4; six7-MO1, n = 6, each dose), One-way ANOVA with Tukey?s post-hoc test. a vs b, p<0.05, b vs c, p<0.001, a vs c, p<0.0001. Error bars represent SD. nb, neuroblast; vp, ventral patch; onl, outer nuclear layer; MO1, morpholino 1. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Photoreceptor progenitor proliferation is regulated by six7. (A) EdU (red) and six7-in situ labeling (blue) in a retinal cryosection from 48-hpf WT embryos. Note the expression of six7 is coincident with proliferating cells in the ONL. (B) EdU labeling in retinal cryosections of WT, six7-MO control and six7 morphants at 52 hpf. Note that EdU labeled cells persist in the central retina in six7 morphants. (C) Retinal cryosections of WT and six7 morphants labeled with EdU at 48 hpf and immunolabeled for rods (4C12, green) at 4 dpf. Proliferative cells are restricted to the ONL and differentiate as photoreceptors in WT and six7-knockdown retinas. Proliferative cells colabeled for rod markers in the central retina, (arrowheads, inset). (D) Retinal cryosections of WT and six7-morphants immunolabeled for phospho-histone 3 (PH3) at 48 hpf and (E) graph showing the number of PH3-positive cells in the INL and ONL per sections (WT, n = 5; and six7-MO1, n = 4; 3?4 sections/retina). Mitosis levels increased significantly in the ONL in six7-morphants. No significant changes were observed in the INL. (***p = 0.0007, Student?s t test). (F) Retinal cryosections of WT and ljrp23ahub at 3 dpf co-labeled for TUNEL (red) and rods (4C12, green), nuclei counterstained with SYTO61. (G) Quantification of TUNEL positive cells in the ONL, INL and GCL of WT (n = 7) and ljrp23ahub (n = 9); (*p<0.05, *****p<0.0001, Student?s t test calculated on log transformed data). (H) Transverse retinal sections from WT and six7-MO1 injected animals at 3-dpf co-labeled for TUNEL (red) and rods (4C12, green), nuclei counterstained with SYTO61. (I) Quantification of TUNEL positive cells in the ONL, INL and GCL of control (n = 7) and six7-MO1 injected embryos (n = 6); (*p<0.05, Student?s t test calculated on log transformed data). Error bars represent SD. Dorsal is up. ONL, outer nuclear layer; INL, inner nuclear layer; GCL, ganglion cell layer; CMZ, ciliary marginal zone. |
|
TALENs-mediated knockout of six7 locus recapitulates ljrp23ahub phenotype. (A) Schematic representation of the Six7 protein domains. six7-TALENs target site and the HaeIII restriction enzyme site are highlighted. The left and right monomer binding sites are underlined. (B) RFLP of six7 locus by HaeIII. A new 170 bp DNA-fragment is detected in six7fl4 carriers compared with controls. (C) Sequence analysis of the six7 target region shows recovery of multiple indel alleles of six7. The target region is highlighted in red. Dots represent deletions and lower case letters indicate insertions. The six7 sequence from WT is shown as a comparison. (D) Confocal immunofluorescent images labeled for rods (red) from WT and six7fl4 knockout mutants at 4 dpf. six7fl4 knockout mutants phenocopy ljrp23ahub mutants. Sequencing chromatograms of WT and six7fl4 mutants illustrated the c.217_229del CAGGTGGCCCGAG (del13) six7 mutation in homozygous zebrafish. The bar graph shows the average number of rods counted in 1?3 different areas per retina (WT (n = 8), six7fl4 (n = 7), six7 fl4/p23ahub (n = 4); One-way ANOVA with Tukey?s post-hoc test. a vs b, p<0.0001. (E) Tangential views of confocal immunofluorescent images labeled for red-sensitive (brighter signal) and green-sensitive (dimmer signal) cones from WT, six7p23ahub, and six7fl4 retinas at 4 dpf. Graph showing the average number of red/green-sensitive cones per unit area of WT (n = 3), six7p23ahub (n = 4), and six7fl4 (n = 3); (ns p> 0.05; a vs b p ? 0.05; significant difference one-way ANOVA with Tukey?s post-hoc test). (F) Whole mount in situ hybridization for RH2 probe (green-sensitive cone opsin) in WT (n = 30), six7p23ahub (n = 30) and six7fl4 embryos (n = 30) at 4 dpf. WT and six7p23ahub embryos often showed the same pattern and number of green-sensitive opsin cone labeling (Dorsal is up and nasal to the left), while six7fl4 knockout showed no labeling or few cells labeling for green-sensitive cone opsins (inset). Graph showing the percentage of unlabeled and labeled embryos. Notice that 17% of the six7p23ahub embryos were un-labeled for green-sensitive cone opsin. (G) Retinal cryosections of in situ hybridization for green-sensitive cone opsin in six7p23ahub (n = 5) embryos immunolabeled with 1D1 (rods). Rods and green-sensitive cone opsin probes labeled different cells in the ONL of six7p23ahub embryos. (H) Evidence of cell death. Flat mount confocal image of nuclei counterstained with DAPI and retinal cryosections from WT and six7fl4 animals at 4dpf co-labeled for TUNEL (red) and rods (4C12, green). six7fl4 mutants (n = 6, 1?2 sections/retina) showed an increase in apoptotic cells, especially in the ONL compared with WT (n = 3, 1?2 sections/retina), (arrows pointing to apoptotic cells in the ONL). |
|
six7 acts cell autonomously. (A) six7fl4-donor cells or (B) WT-donor cells were labeled by the tracer rhodamine-dextran, transplanted into WT genetic background and allowed to develop until 4 dpf. Expression of rods (4C12, green) and green-sensitive opsin (pink) were detected by whole mount immunolabeling. Note that six7fl4-transplanted cells frequently differentiate as rod photoreceptors (orange, arrows) and rarely immunolabeled for green-sensitive opsins while neighboring WT cells differentiate as green-sensitive cones. Significant difference was observed in the percent of six7fl4-donor cells that differentiate into rods or cones compared to WT donor cells (?2, p<0.0001). |
|
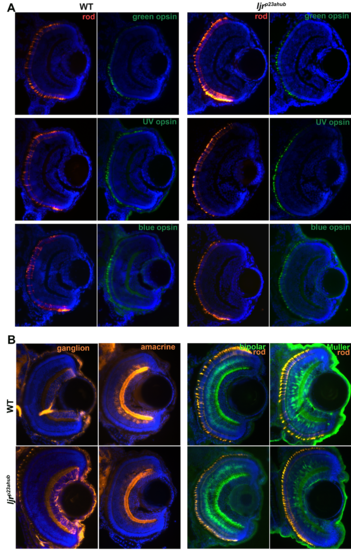
(A) No changes in green-, UV- and blue-sensitive cone opsin expression in ljrp23ahub mutants. Retinal cryosections from WT and ljrp23ahub embryos at 4 dpf immunolabeled for rods (4C12, red) and the green-, UV- and blue-sensitive cone opsins (green). Nuclei were counterstained with DAPI (blue); dorsal is up. No differences in the number or expression levels are detected for any of the opsin subtypes, except for the increased number of rods in ljrp23ahub mutants. (B) retinal cryosections from WT and ljrp23ahub embryos at 4 dpf immunolabeled for ganglion cells (Zn8), amacrine cell (5E11), bipolar cells (PKC?) and rods (4C12). No differences were observed for any labeled retinal cell types. |
|
six3a/b expression is unchanged in six7 morphants. (A) Linkage analysis places the ljrp23ahub locus on chromosome 7, 6 out of 463 larvae show recombination at marker ZC168D1-SSR6 and 1 out of 252 larvae show recombination at marker ZC42C5-SSR2. (B) Diagram of predicted morpholino recognition sites (bars) in six7 loci. MO1 targets a translational site and MO3 blocks a donor splice site in intron 1 of six7. Incorrect splicing can be seen in morpholino-injected animals using primers in exon1 and 2 (arrows). (C) RT-PCR fragments, using primers highlighted in A, were analyzed by 1% agarose gel electrophoresis. Arrow highlights the six7 alternative spliced product obtained at 12 hpf (* indicates the new cryptic splice site). (D) No changes in the expression of the homologues six3a (left graph) or six3b (right graph) were detected between WT and ljrp23ahub mutants (n = 30 embryos per group). All the real-time PCR experiments were carried out in triplicates and normalized to ?-actin. (E) Plastic sections of 4 dpf retinas from WT and six7-MO1 injected larvae. The three layers are regularly arranged in WT and morphant retinas. Close examination of the retina revealed a densely packed ONL in morphants. Graph showing the average number of nuclei per unit area (WT, n = 3, 2 sections each; six7-MO1, n = 3, 2 sections each). Cells are increased in the ONL of six7 morphants. Student t test, arcsine transformation, *p<0.05. (F) Electron micrographs of rod outer segments (ROS) from central retina of WT and six7-MO1 larvae. ROS from six7 morphants show their typical morphology of parallel- flattened sacs with continuously membranes at the edges (arrows) indistinguishable from WT ROS. |
|
six7 functions in photoreceptor progenitor cells. (A) Histological sections of chimera retinas labeled for rods (4C12, green). six7-MO donor cells (red) preferentially generate rods compare to WT donor cells. Note the gap in rod labeling in WT/WT controls. Graph represents the percentage of donor cells that differentiate as rods in the central retina of WT/WT (n = 5) and six7-MO1/WTchimeras (n = 6). *p<0.05, student t test. (B) rx1-, pax6a-, crx- and neurod-in situ hybridization (blue) in a retinal cryosection from 48-hpf WT and six7-MO1 embryos. No labeling of retinal progenitors cell markers (rx1 and pax6a) were observed in the ONL of six7-morphant retinas. (C) Whole-mount in situ hybridization for the dorsal (tbx2b), midline (cyp26c1) and ventral (vax2) retinal marker. Labeling was indistinguishable different between WT and mutant embryos. |
|
Levels of mitosis are altered in six7fl4 at 56 hpf. (A) Confocal immunofluorescent images labeled for rods (4C12, red) from WT, six7fl4 and six7fl4/p23ahub retinas at 4 dpf. (B) Retinal cryosections from carrier animals (six7fl4/+, n = 5, 1?2 sections/retina) and six7 (n = 11) embryos at 56 hpf co-labeled for TUNEL (red) and PH3 (green), nuclei counterstained with DAPI. No differences in TUNEL labeling were detected. Graphs showing the number of PH3+ cells by section (excluding CMZ). Number of PH3+ cells is significantly greater in ONL of six7fl4 mutants at 56 hpf. Un-paired Student t test with Welch?s correction, *p<0.05). (C) Retinal cryosections from WT and six7fl4 embryos at 4 dpf immunolabeled for rods (4C12, red) and the green-, UV-, blue- and red-sensitive opsins (green). Nuclei were counterstained with DAPI (blue). Dorsal is up. Depleted green-sensitive opsin expression is noticeable in six7fl4, but other opsins appear unaltered. |