- Title
-
The bHLH transcription factor Ascl1a is essential for the specification of the intestinal secretory cells and mediates Notch signaling in the zebrafish intestine
- Authors
- Flasse, L., Stern, D.G., Pirson, J.L., Manfroid, I., Peers, B., and Voz, M.L.
- Source
- Full text @ Dev. Biol.
|
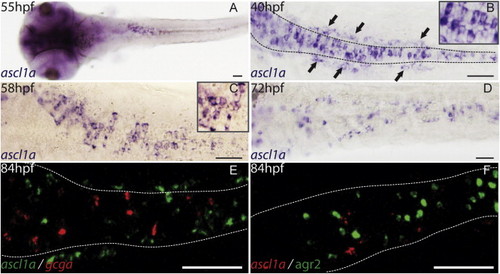
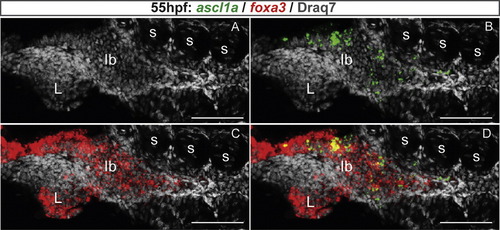
ascl1a is expressed in the precursor cells of the gastrointestinal tract. Ventral views with anterior on the left of embryos analyzed by visible WISH (A?D) or double fluorescent WISH (E and F). (A) General view of ascl1a expression domain at 55 hpf. (B) At 40 hpf, ascl1a is expressed in cubo´dal cells scattered in the primitive intestine (dotted lines) and in enteric neurones that lined these cells (arrows). (C) At 58 hpf, this scattered expression is still intense. (D) At 72 hpf, the number of ascl1a expressing cells begins to decrease along the gastrointestinal tract. (E and F) Confocal sections showing that ascl1a is not expressed in the mature enteroendocrine cells labeled by the gcga probe (E) neither in the mature goblet cells labeled by agr2 (F). Scale bars: 50 μM. EXPRESSION / LABELING:
|
|
ascl1a is expressed in the precursor cells of the gastrointestinal tract. Ventral views with anterior on the left of embryos analyzed by visible WISH (A?D) or double fluorescent WISH (E and F). (A) General view of ascl1a expression domain at 55 hpf. (B) At 40 hpf, ascl1a is expressed in cuboïdal cells scattered in the primitive intestine (dotted lines) and in enteric neurones that lined these cells (arrows). (C) At 58 hpf, this scattered expression is still intense. (D) At 72 hpf, the number of ascl1a expressing cells begins to decrease along the gastrointestinal tract. (E and F) Confocal sections showing that ascl1a is not expressed in the mature enteroendocrine cells labeled by the gcga probe (E) neither in the mature goblet cells labeled by agr2 (F). Scale bars: 50 μM. EXPRESSION / LABELING:
|
|
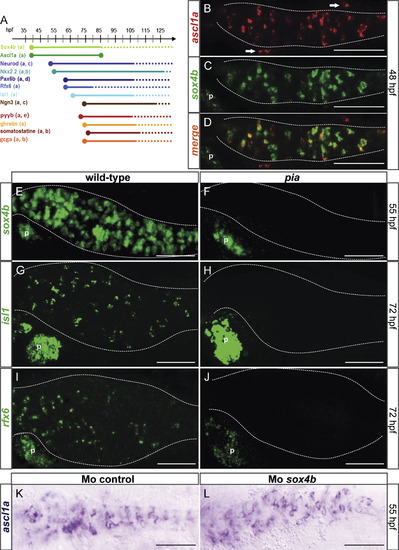
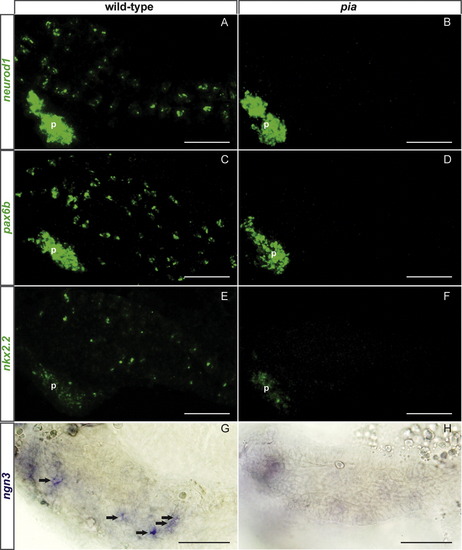
Ascl1a is a key regulator of the secretory cell differentiation cascade. (A) Diagram illustrating the expression window of the transcription factors and hormones in the gastrointestinal tract. The interval of time where the gene has been shown to be expressed is represented by a solid line whereas the gene expression was not analyzed during the periods outlined by dotted lines. The references for these data are as follows: a (this paper), b (Ng et al., 2005), c (Zecchin et al., 2007), d (Delporte et al., 2008), e (Wallace et al., 2005). (B?L) Ventral views with anterior to the left of embryos analyzed by fluorescent WISH (B?J) or visible WISH (K and L) (B?D) Confocal sections showing that ascl1a expression co-localize perfectly with sox4b expression in scattered cells of the primitive intestine (dotted line) at 48 hpf. (E?J) Confocal projections showing that all the gastrointestinal cells expressing sox4b, isl1 or rfx6 are absent in the pia mutant whereas the pancreatic expression of these factors is not affected (K and L) ascl1a expression is not affected upon Sox4b knock-down. P, pancreas; Arrow, enteric neurone. Scale bars: 50 μM. EXPRESSION / LABELING:
|
|
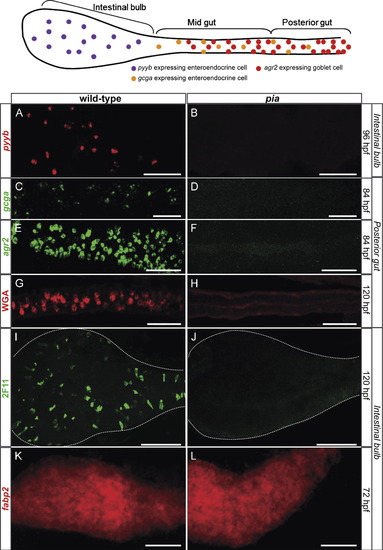
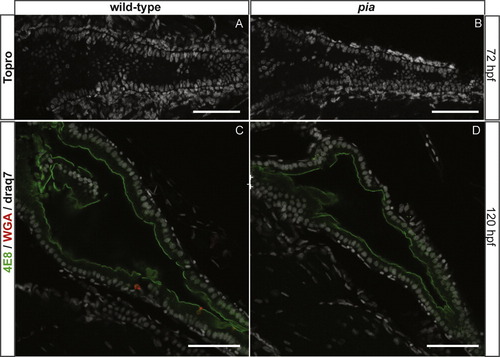
All the cells of the gut epithelium adopt either a goblet or enteroendocrine fate in the mib mutant. Ventral views with anterior to the left of wt or mib embryos labeled by immunohistochemistry with 2F11 together with the binding of fluorescent WGA (A and B) or by WISH using the agr2 probe (C and D) or the fabp2 probe (E and F). A and B: The gut of 5-day mib mutants is nearly completely covered by secretory cells, either enteroendocrine cells (2F11+/WGA?), found preferentially in the intestinal bulb and the midgut, or goblet cells (2F11+/WGA+). C and D: The presence of goblet cells in the gut of mib mutant at 4 dpf was confirmed by WISH using the agr2 probe. The double-headed arrows indicate the length of the gut. E and F: 84 hpf mib larvae display a nearly complete loss of fabp2-labeled enterocytes. Scale bars: 50 μM. EXPRESSION / LABELING:
|
|
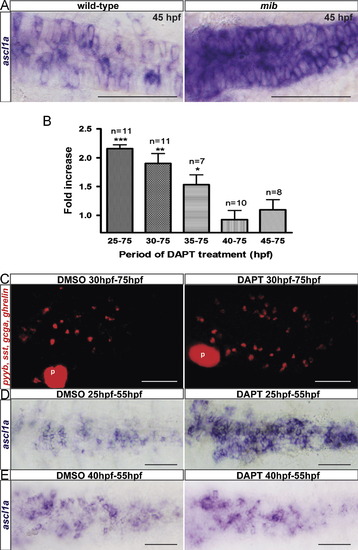
Notch signaling acts between 25 hfp and 40 hpf to repress enteroendocrine cell differentiation and ascl1a expression. Ventral views with anterior to the left of embryos analyzed by visible (A, D and E) or fluorescent (C) WISH. (A) ascl1a expression is strongly increased in the primitive gut of 45 hpf mib embryos. (B) Quantification of the total number of enteroendocrine cells present in the whole intestine and labeled with a mixture of pyyb, gcga, sst and ghrelin. These positive cells have been directly counted under the microscope. Fold increase represents the mean (+S.EM.) of the enteroendocrine cells number in DAPT treated embryos versus DMSO treated embryos. Asterisks indicate that the difference between the cell number in control and embryos treated with DAPT is statistically significant by Student′s t-test. ***P<0.0001; **P<0.001; *P<0.05. (C) Confocal projections of 75 hpf embryos labeled with a mixture of pyyb, gcga, sst and ghrelin probes and treated with DAPT or DMSO from 30 hpf to 75 hpf. P, pancreas (D and E) ascl1a expression at 55 hpf is increased in the primitive gut of embryos treated with DAPT from 25 to 55 hpf (E) but not in embryos treated from 40 to 55 hpf (G). |
|
Ascl1a is required for the induction of secretory cell fate upon inhibition of the Notch pathway. Ventral views with anterior to the left of embryos analyzed by double fluorescent WISH using a agr2 probe (green) and with a mixture of gcga, sst and ghrelin probes (red). All pictures are confocal projections. (A and C) WT embryos treated with DAPT from 25 to 96 hpf show an increased number of enteroendocrine and goblet cells (fold increase of 2.2▒0.5 and 1.5▒0.4, respectively). (B and D) pia embryos treated with DMSO or DAPT from 25 to 96 hpf are devoid or secretory cells in the gastrointestinal tract. P, pancreas. Scale bars: 50 μM. EXPRESSION / LABELING:
|
|
|
|
|
|
|
Reprinted from Developmental Biology, 376(2), Flasse, L., Stern, D.G., Pirson, J.L., Manfroid, I., Peers, B., and Voz, M.L., The bHLH transcription factor Ascl1a is essential for the specification of the intestinal secretory cells and mediates Notch signaling in the zebrafish intestine, 187-197, Copyright (2013) with permission from Elsevier. Full text @ Dev. Biol.