- Title
-
Mutations in zebrafish leucine-rich repeat-containing six-like affect cilia motility and result in pronephric cysts, but have variable effects on left-right patterning
- Authors
- Serluca, F.C., Xu, B., Okabe, N., Baker, K., Lin, S.Y., Sullivan-Brown, J., Konieczkowski, D.J., Jaffe, K.M., Bradner, J.M., Fishman, M.C., and Burdine, R.D.
- Source
- Full text @ Development
|
seahorse encodes Lrrc6l. (A-A″) sea encodes a 440 amino acid protein (Ai); the N terminus contains four LRR motifs terminated by an LRR cap motif (green; Aii). The fourth LRR motif is disrupted by the seatg238a L93P missense mutation (pink). seafa20r Q201X nonsense mutation truncates the protein shortly after the coiled-coil (pink). The protein is predicted to contain three weak nuclear export signals (NES) and one strong nuclear localization signal (Aiii). (A′) Lrrc6l is conserved among species. An ortholog from T. brucei is 39% identical to amino acids 1-363 of Sea. Orthologs from Xenopus and mouse are 55% identical and 54% to amino acids 1-440 of Lrrc6l, respectively, and extend roughly 30 amino acids beyond the Lrrc6l C terminus. (A″) Alignment of the four Lrrc6l LRR motifs; the consensus Lrrc6l LRR sequence is [L/C]xxLxxLxLxxNxIxxIxxVxx(x) and is most similar to repeats in the SDS22-like subfamily (Kobe and Kajava, 2001). The core LRR consensus sequence shared among all LRR proteins, LxxLxLxxN/CxL (x can be any amino acid and L positions can be V, I or F), is underlined (Kobe and Kajava, 2001). (B-B″′) seafa20r is a 702C→T transition in exon 5 encoding a Q201X truncation (B,B′). seatg238a is a 273T→C transition in exon 3 encoding an L93P missense (B″,B″′). (C-C″′) Genotyping (C,C″) seafa20r (red) creates an AluI restriction site (black nucleotides), cleaving the 114 base pair (bp) PCR product into 81 and 33 bp fragments. (C′,C″′) seatg238a (red) abolishes an FspBI restriction site (black nucleotides), preventing cleavage of the 176 bp fragment into 114 and 62 bp fragments. (D,D′) Genotyping results. The left-most lanes contain DNA size markers with upper band at 200 bp and lower band at 100 bp. |
|
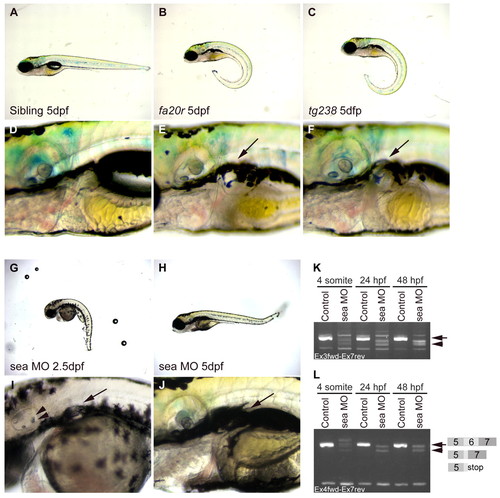
seahorse mutants and sea MO-injected embryos displayed curly tail down and pronephric cyst phenotypes. (A-F) CTD and pronephric cyst phenotypes in sea mutants. (A) Siblings, (B) seafa20r and (C) seatg238a; mutant embryos have CTD phenotypes at 5 dpf. (D) Siblings, (E) seafa20r and (F) seatg238a embryos at higher magnification to show pronephric cysts at 5 dpf (arrows). (G-J) CTD and pronephric cyst phenotypes in sea MO-injected embryos. MO-injected embryos can recover from the CTD (H) and cyst phenotypes (J) by 5 dpf. Defects in otic vesicle and otolith formation (arrowheads) were observed in some sea MO-injected embryos (I). (K,L) RT-PCR from uninjected and sea MO-injected cDNA libraries at four somites, 24 hpf and 48 hpf. (K) Primers between exon 3 and exon 7 amplified a wild-type sea band at 635 base pairs (bp; arrow) and incorrectly spliced message in sea MO-injected embryos (arrowhead). (L) Primers between exon 4 and exon 7 amplified a wild-type sea band at 517 bp (arrow; exon 5-7) and incorrectly spliced message in sea MO-injected embryos (arrowhead). Incorrect splicing generated two main splice forms creating either a deletion and/or stop codon in exon 6 (indicated by the diagram in L). EXPRESSION / LABELING:
|
|
seahorse mRNA is expressed in tissues that possess motile cilia. (A) RT-PCR of sea mRNA (503 bp) in cDNA libraries at the one- to two-cell, 256-cell, 1024-cell, sphere and 18-somite stages. sea mRNA was maternally expressed as zygotic transcription initiates after the 1024-cell stage. (B-J) Expression of sea was detected by RNA in situ hybridization in dorsal forerunner cells at 90% epiboly (B), in KV at 3 somites (C), and in the floor plate at 22 somites (D, arrowheads), 24 hpf (E,F arrowheads; G, bracket) and 48 hpf (J, arrowhead). Expression was detected in the pronephric tubules at 22 somites (D, arrows), 24 hpf (E,F, arrows), 36 hpf (H, arrow) and at 48 hpf (I,J, arrow). Open arrows in G and I indicate sea expression in the chordoneural hinge. Double-headed arrows in D,F and G span the width of the notochord, sea mRNA is not detected. Inset in F is a higher magnification image of the patchy pronephric expression that resembles expression of genes in multiciliated cells (Liu et al., 2007; Ma and Jiang, 2007). |
|
Mutations in lrr6l do not affect general pronephric patterning or cilia structure. (A-D) Expression of pronephric specification genes at eight somites is not affected in sea mutants (B,D) compared with wild type (A,C). (A,B) wt1 is correctly expressed in the intermediate mesoderm from the first somite to the beginning of the fourth somite. (C,D) pax2.1 is correctly expressed in the intermediate mesoderm posterior to the third somite. In A-D, RNA in situ hybridization for α-tropomyosin was used to visualize the somites. (E,F) Expression of pax2.1 in the neck segments at 48 hpf is unaffected in sea mutants (F) compared with siblings (E). (G,H) Acetylated tubulin immunofluorescence of KV at six somites in sea mutant (H) and sibling (G) shows that cilia are not affected in sea mutants. (I-K) In wild-type embryos at 72 hpf (I), wt1 is expressed in the fused glomerulus in the midline. In sea mutants, defects in wt1 expression were seen, including separation of the glomeruli (J) and drastic expansions of the wt1 domain (K). These effects are probably caused by the expansion of the glomerular region. (L,M) Acetylated tubulin staining in sibling (L) and sea mutants (M) at 27 hpf indicate that cilia formation in the pronephros is unaffected by mutations in sea. All above images are from seafa20rmutants, but similar results were obtained for seatg238a. (N-Q) TEM analysis of pronephric cilia show no alterations in axoneme structure in seafa20r mutants (O,P) compared with siblings (N). Basal body localization at the apical membrane is also unaffected in sea mutants (Q; arrow indicates basal body). Anterior is towards the top in A-K. L and M are lateral views of the pronephric tubules; anterior is towards the left. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Pronephric cyst phenotypes in seahorse mutants and sea MO-injected embryos. (A-F) At 2 dpf, the glomerular region (asterisks) appears normal in siblings (A) and seafa20r mutants (B), and slightly dilated in sea MO-injected embryos (C). In the same embryos, the medial tubules (arrows) were significantly dilated both in seafa20r mutants (E) and sea MO-injected embryos (F), compared with siblings (D). (G-L) At 2.5 dpf, the glomeruli (asterisks) in both the seafa20r mutants (H) and sea MO-injected embryos (I) were dilated compared with siblings (G). These same embryos have dilations in the medial tubules (arrows) in seafa20r mutants (K) and sea MO-injected embryos (L) compared with siblings (J). (M-O) At 3 dpf, siblings (M) display basolateral localization of Na+/K+-ATPase (red) in the pronephric epithelium (arrows; white arrowheads indicate lateral staining), whereas the sea mutants (N,O) display altered localization of Na+/K+-ATPase diffusely at apical (yellow arrowheads) and lateral membranes (white arrowheads). EXPRESSION / LABELING:
PHENOTYPE:
|
|
seahorse mutants retain the ability to interact with Dsh. Upper panel: western blot with anti-HA on samples pulled down by immunoprecipiatation with anti-Flag. Middle and lower panels are western blots of lysates with the indicated antibody to visualize the input of Sea proteins (HA) or Dsh (Flag). Note that both seatg238a and seafa20r can produce stable proteins in vivo. Both mutant Sea proteins can interact with Dsh, although the weaker interaction seen with Seafa20r may indicate that the truncation in this protein affects the binding site for Dsh. |
|
seahorse mutants have a low level of left-right patterning defects. (A) At 20 somites, the majority of sea mutants have normal left-sided expression of southpaw and lefty1/2 in the lateral plate mesoderm, and lefty1 in the diencephalon (asterisk). southpaw is expressed lateral (marked with arrows) to the darker stained lefty1/2-expressing cells in the lateral plate. (B-G) The majority of sea mutants had correctly looped hearts (B) with the ventricle (white V) looped to the right of the atrium (white A) and correctly placed visceral organs (E) with the liver (white L) on the left and pancreas (white P) on the right. A low level of sea mutants display either situs inversus (complete reversal of organ positioning; C,F) or heterotaxia (random organ positioning; D,G). (H-J) sea mutants also display a low level of asymmetric defects in habenular expression of lov at 4 dpf. Although most mutants have the typical greater level of expression on the left (H), some mutants have reversed (I) or isomeric (J) expression. (A) Dorsal view, anterior towards the top. (B-D) Ventral views, anterior towards the top. (E-G) Dorsal views of the same embryos in B-D, anterior towards the top. (H-J) Dorsal views of the diencephalon, anterior towards the top. In all panels, black R indicates right, black L indicates left. EXPRESSION / LABELING:
PHENOTYPE:
|

Unillustrated author statements |