- Title
-
Pbx homeodomain proteins direct Myod activity to promote fast-muscle differentiation
- Authors
- Maves, L., Waskiewicz, A.J., Paul, B., Cao, Y., Tyler, A., Moens, C.B., and Tapscott, S.J.
- Source
- Full text @ Development
|
Pbx and Myod are required for the proper initiation of myog expression. (A-R) RNA in situ expression of (A-C,G-I) myod and krox-20, (D-F,J-L) myog and krox-20, (M-O) myog (blue) and myod (red), or (P-R) desm and krox-20 in (A,D,G,J,M,P) wild-type control, (B,E,H,K,N,Q) pbx2-MO; pbx4-MO, (C,F,I,L,R) myod-splMO, or (O) myod-MO embryos. Similar phenotypes were observed for both myod splice-blocking MOs (myod-splMOs). Somite (s) staging was confirmed by counting somites using Nomarski optics. All embryos are shown in dorsal view, anterior towards the left. (A) Arrowheads point to adaxial cells; arrows point to lateral somite cells; and r3 and r5 indicate krox-20 expression in hindbrain rhombomeres. (S-U) Graphs of real-time reverse-transcriptase (RT)-PCR quantities for (S) myod, (T) myog and (U) desm. (T, bottom) Enlarged view of 6s stage myog expression and the key for S-U. x-axes are developmental stage and y-axes are relative mRNA expression level. Quantities were normalized to odc1 expression. Error bars represent standard deviation. myodE2I2-splMO targets the exon 2-intron 2 boundary (see Fig. S3 in the supplementary material). The myod real-time primers measure only mRNA with properly spliced intron 2. Quantitative (q)RT-PCR shows that less than about 10-15% of normally spliced myod transcripts remain in myod-splMO embryos (Fig. 1S and see Fig. S3 in the supplementary material; and data not shown). Similar quantities for myog and desmin were observed for both myod-splMOs. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Validation of microarray-identified Pbx-dependent genes. (A-Z) RNA in situ expression of genes from Table 1 in (A,C,E,G,I,K,M,O,Q,S,U,X) wild-type control, (B,D,F,H,J,L,N,P,R,T,V,Y) pbx2-MO; pbx4-MO, or (W,Z) myod-MO embryos. (S,T) Control gene, desm; (X-Z) slow-muscle gene, smyhc1. Although pbx2-MO; pbx4-MO embryos appear delayed relative to controls, all embryos are stage-matched at (A-T) 18-somite stage (18s), (U-W) 16s or (X-Z) 14s. krox-20 was included in all in situs to control for the pbx MOs. (A) r3 and r5 indicate krox-20 expression in hindbrain rhombomeres. (Q) Arrow marks vmhc expression in the heart primordium. Embryos are shown in (A-P,S-T) left-side view or (Q-R,U-Z) dorsal view, anterior towards the left. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Pbx is required for the proper initiation of fast-muscle differentiation. (A-D) Antibody staining of slow muscle with F59 antibody (green; A,B), or fast muscle with F310 antibody (green; C,D), in (A,C) wild-type control or (B,D) pbx2-MO; pbx4-MO embryos. Antibody staining of muscle with MF20 antibody (red; A-D) is included to control for somite staging. Embryos are shown in left-side view, anterior towards the left. (E,F) Quantification of (E) F59/slow- and (F) F310/fast-muscle marker expression. MF20 expression is included to control for somite (s) staging. Graphs show number of somites expressing a muscle marker at different developmental stages. Similar phenotypes were observed for both myod splice-blocking MOs (myod-splMOs). Error bars represent standard deviation. P values (Mann-Whitney test; normalized to somite number): *P<0.0001 compared to same-stage control; except for pbx2-MO; pbx4-MO F310 compared to control at 24 hpf, P<0.03. PHENOTYPE:
|
|
Pbx functions with Myod in fast-muscle, but not slow-muscle, differentiation. RNA in situ expression at the 10-somite (10s) stage of (A-G) myog and krox-20, (H-N) desm and krox-20 or (O-U) smyhc1 and krox-20, or at 16s of (V-BB) mylz2 and krox-20 in (A,H,O,V) wild-type control, (B,I,P,W) pbx2-MO; pbx4-MO, (C,J,Q,X) myod-MO, (D,K,R,Y) myf5-MO, (E,L,S,Z) myf5-MO; myod-MO, (F,M,T,AA) myf5-MO; pbx2-MO; pbx4-MO, or (G,N,U,BB) myod-MO; pbx2-MO; pbx4-MO embryos. Embryos are shown in dorsal view, anterior towards the left. Somite staging was confirmed by counting somites using Nomarski optics. (A) r3 and r5 indicate krox-20 expression in hindbrain rhombomeres. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Pbx is required downstream of Shh signaling to induce fast-muscle, but not slow-muscle, gene expression. (A-L) RNA in situ expression of (A-F) smyhc1 (blue) and myod (red) at the 14-somite (14s) stage or (G-L) mylz2 (blue) and myod (red) at 16s in embryos injected with (A,G) pbx2-MO mismatch control and gfp mRNA, (B,H) pbx2-MO mismatch control and shh mRNA, (C,I) myod-MO and gfp mRNA, (D,J) myod-MO and shh mRNA, (E,K) pbx2-MO; pbx4-MO and gfp mRNA, or (F,L) pbx2-MO; pbx4-MO and shh mRNA. The most-anterior 8-10 somites of each embryo are shown, in dorsal view, anterior towards the left. Control embryos injected with shh mRNA (control+shh) show increased expression of smyhc1 (B, 106/135 embryos; 29/135 show normal smyhc1 expression) or increased expression of mylz2 (H, 88/121; 33/121 show normal mylz2 expression). myod-MO+shh embryos show less-frequent and less-extensive induction of smyhc1 compared with control+shh (46/90 embryos resemble D; 44/90 show normal smyhc1 expression) and fail to upregulate mylz2 expression beyond that seen in myod-MO+gfp embryos (84/88 embryos resemble J; 4/88 show slightly increased mylz2 expression). pbx2-MO; pbx4-MO+shh embryos show strongly increased expression of smyhc1 (77/79 embryos resemble F; 2/79 have normal smyhc1 expression). Approximately half (43/102; L) of pbx2-MO; pbx4-MO+shh embryos show expansion of weak levels of mylz2 across the somites; the other half (59/102) resemble pbx2-MO; pbx4-MO+gfp embryos. Similar results were observed in multiple experiments. |
|
Pbx expression during early myogenesis stages is lost in pbx2-MO; pbx4-MO embryos. (A-D) Anti-zebrafish pan-Pbx staining in (A-C) whole wild-type control and (D) pbx2-MO; pbx4-MO embryos at (A) bud stage/10 hpf, (B) 10 somite stage (s)/14 hpf, or (C) 24 hpf. 24 hpf embryos were also stained with Sytox Green to identify nuclei. (A,B) Show dorsal views, anterior towards the left, of (A) early paraxial mesoderm and (B) somites 3-5. (A) Arrowheads label the rows of adaxial cells next to the notochord. (C,D) Show lateral views, anterior towards the left, centered on somites 8-11. (E) Western blot using zebrafish pan-Pbx antibody and Histone H4 antibody. Western analysis with a Pbx4-specific antibody identified the two Pbx4 bands (data not shown). |
|
pbx2-MO; pbx4-MO embryos are rescued by exd. (A-D) RNA in situ expression of myog and krox-20 (blue) and myod (red). A and B are wild-type control siblings of C and D, which were injected with pbx2-MO; pbx4-MO. A and C were injected with gfp mRNA, and B and D were injected with exd mRNA. In this experiment, embryos were fixed at the same time, about 10s-stage for A,B,D and about 8s-stage for C. exd mRNA rescues krox-20 and myog expression in pbx2-MO; pbx4-MO embryos. exd mRNA also rescues the developmental delay (scored by myod expression) in pbx2-MO; pbx4-MO embryos. Embryos are shown in dorsal view, anterior towards the left. (E) Quantification of the rescue experiment by counting the number of somites expressing myog or myod. Error bars represent standard deviation. |
|
myod splice-blocking and translation-blocking morpholinos knock down normal myod mRNA or protein levels. (A) Diagrams of functional domains of Myod, including the acidic activation domain, and the histidine- and cysteine-rich domain (H/C). The amino acid sequence of helix III is 100% identical between zebrafish, mouse and human. Exon 1-exon 2 (E1/E2) and exon 2-exon 3 (E2/E3) junctions are shown. (B) RT-PCR analysis of myod mRNA products (using primers in exon 1 and exon 3) from wild-type control, myod-splMO E1I1 and myod-splMO E2I2 embryos. E1I1 targets the myod exon 1-intron 1 boundary and E2I2 targets the exon 2-intron 2 boundary. cDNA sequencing confirms that the E1I1 splice variant retains intron 1 and the E2I2 splice variants retain all (variant a) or most (variant b) of intron 2. qRT-PCR shows that less than about 10-15% of normally spliced myod transcripts remain in myod-splMO embryos (Figs 2P and S3 and data not shown), whereas the total level of myod mRNA remains similar or increased compared to that in control embryos (Fig. S3 and data not shown). (C) Predicted amino acid sequences based on cDNA sequencing of splice variants. Helix III domain is underlined; amino acid sequence from translated intron sequences are shown in italics. When intron 1 or intron 2 splicing is inhibited, in-frame stop codons in intron 1 or intron 2 predict the generation of truncated Myod proteins lacking helix III. E2I2 variants a and b share the same intron sequence up to the stop codon. (D-G) Mammalian anti-Myf5 antibody (which detects zebrafish Myod protein; Hammond et al., 2007) staining in (D) control, (E) myod-MO, (F) myod-splMO E1I1, and (G) myod-splMO E2I2 embryos. The anti-Myf5 antibody was generated against the C-terminus of the protein. Therefore, although myod-splMO embryos appear to have little or no wild-type Myod protein, we cannot determine whether the predicted truncated product is present. EXPRESSION / LABELING:
|
|
Fast-muscle differentiation is delayed and reduced in pbx2-MO; pbx4-MO and in myod-MO embryos. (A-C) Antibody staining of fast-muscle myosins with F310 (green in A-C) or pan-muscle myosins with MF20 (red in A-C) in (A) wild-type control, (B) pbx2-MO; pbx4-MO, or (C) myod-MO embryos, all at 16s stage. Somites 4-11 of each embryo are shown in dorsal view, anterior towards the left. MF20 is expressed most strongly in adaxial cells at this stage and appears normal in pbx2-MO; pbx4-MO and in myod-MO embryos. F310 staining is reduced in more posterior somites of pbx2-MO; pbx4-MO and myod-MO embryos. |
|
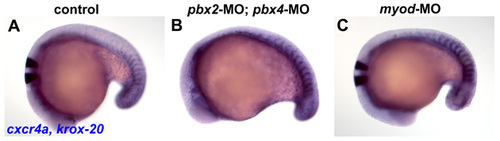
cxcr4a expression is maintained in anterior somites in pbx2-MO; pbx4-MO and in myod-MO embryos. (A-C) RNA in situ hybridization of cxcr4a at 18s in (A) wild-type control, (B) pbx2-MO; pbx4-MO, or (C) myod-MO embryos. cxcr4a is normally expressed in the anterior-lateral cells of only tail somites at this stage (Hollway et al., 2007). Embryos are shown in dorsal view, anterior towards the left. |
|
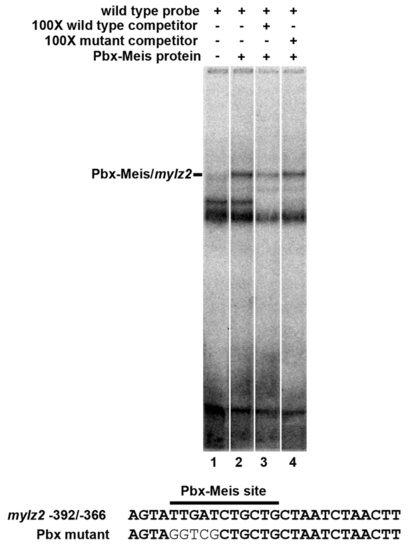
Pbx-Meis heterodimers bind to the zebrafish mylz2 promoter. EMSA using in vitro translated zebrafish Pbx4 and Meis3 proteins and a probe from sequences -392/-366 of the zebrafish mylz2 promoter (Ju et al., 2003). An excess of unlabeled wild-type probe effectively competes the shifted species, whereas a competitor with a mutant Pbx site does not. |