Fig. 3
|
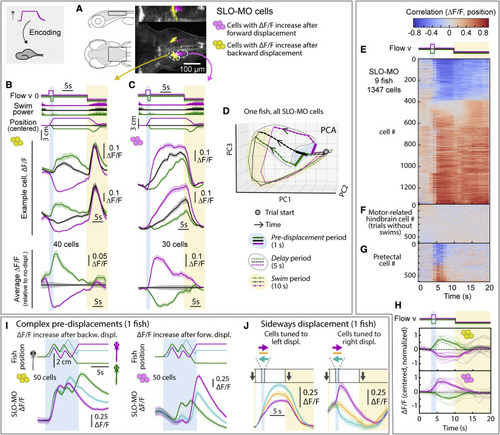
SLO-MO neuronal activity encodes self-location (A) Location of neurons with increasing activity following backward (yellow) versus forward (magenta) pre-displacement in example fish. (B) Examples (top) and average (bottom, with ?F/F during no-displacement trials subtracted) of neurons with increasing activity following backward pre-displacement. (Shaded regions: SEM in all panels; here, 2 s forward or 1 s backward pre-displacements.) (C) Examples (top) and averages (bottom) of neurons with increasing activity following forward pre-displacement. (D) Principal-component analysis embedding of SLO-MO population activity. Trajectories remain separated throughout delay period, gradually converge and return toward the starting point during swim period. (E) SLO-MO neuron activity across 9 fish (1,347 cell segments) sorted by Spearman correlation, showing consistent correlation to self-location across trial types. (F and G) For comparison, lack of correlation to self-location for cells with activity correlating to swim vigor, and for cells in pretectum that show visual encoding. (H) Averages of ?F/F (centered to zero pre-displacement, normalized in each fish) of positive- and negative-correlating neurons from (E). (I) Encoding of self-location by SLO-MO neurons during complex trajectories (example fish). (J) Encoding of sideways changes in self-location in SLO-MO neurons (example fish). |
Reprinted from Cell, 185, Yang, E., Zwart, M.F., James, B., Rubinov, M., Wei, Z., Narayan, S., Vladimirov, N., Mensh, B.D., Fitzgerald, J.E., Ahrens, M.B., A brainstem integrator for self-location memory and positional homeostasis in zebrafish, 50115027.e205011-5027.e20, Copyright (2022) with permission from Elsevier. Full text @ Cell