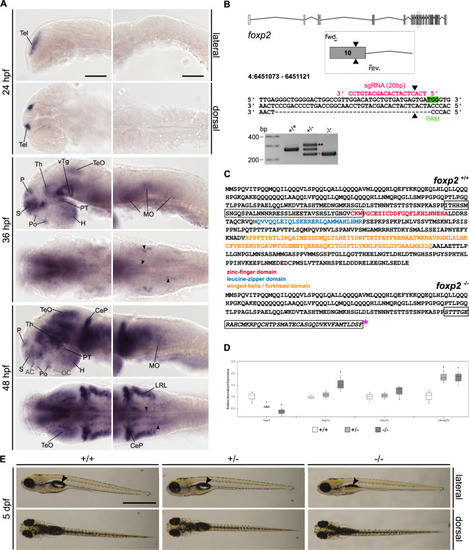
A Whole-mount RNA in situ hybridisation (ISH) in the developing zebrafish. Anterior is to the left. Abbreviations are listed in Table S4. Arrowheads indicate distinct foxp2-positive populations in the MO. Scale bars: 100 µm. B foxp2 exon?intron structure with coding (grey) and non-coding exons (white). The sgRNA (pink) targeting foxp2 exon 10 and the primer binding sites for genotyping PCR are indicated. The CRISPR/Cas9-induced double-strand break (black triangle) caused a 40 bp deletion represented by a PCR product of 228 bp (pink asterisk) in foxp2+/? and foxp2?/?. A third band of ~320 bp corresponds a heterodimer of wildtype and mutated PCR product (black asterisks). C Predicted amino acid sequence of the wildtype (top) and mutated allele (bottom). The frameshifted sequence (black box) is interrupted by a premature stop codon (pink asterisk) N-terminal of the zinc-finger domain. D Relative normalised expression of foxp2, foxp1a, foxp1b and cntnap2b in foxp2+/+ (white), foxp2+/? (light grey) and foxp2?/? (dark grey) based on qPCR. *P < 0.05. E Live images of 5 dpf old foxp2+/+, foxp2+/? and foxp2?/?. Anterior is to the left. foxp2?/? show alterations in the development/inflation of the swim bladder (black arrow) in 20% of the cases. Scale bar, 1 mm.
|

