|
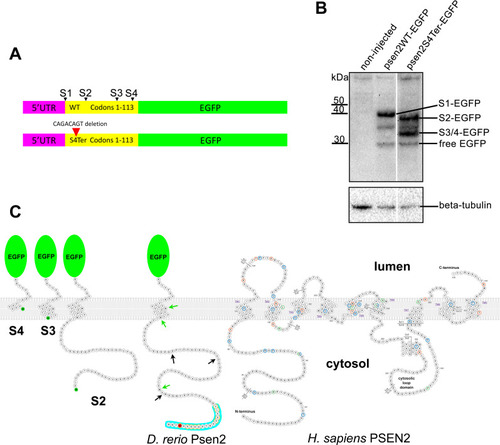
Testing for translation initiation at novel downstream start codons.(A) The constructs, psen2WT-EGFP (upper) and psen2S4Ter-EGFP (lower) used to test for translation initiation at Met codons downstream of the S4Ter mutation. S1 represents the wild type translation start site and S2-4 are potential downstream translation initiation sites within the first 113 codons. (B) Western immunoblotting of lysates from embryos injected with the constructs described in A. Translation initiation at S1 (S1-EGFP) does not permit initiation at S2-4. However, in the presence of the S4Ter mutation, initiation is evident at S2 (S2-EGFP) and either S3 or S4 or both (S3/4-EGFP). The identity of the low intensity ~35kDa protein band is not known but it may be a degradation product of larger fusion protein species. Free EGFP can be produced from EGFP fusion protein species in the lysosome [53, 54]. (C) A representation of the fusion proteins predicted to be produced after injection into embryos of psen2S4Ter-EGFP mRNA and translation initiation at S2, S3, or S4. Note that their representation here as inserting into a lipid bilayer cannot be assumed. Green arrows indicate the positions of downstream alternative possible in-frame start codons (Met codons) in the zebrafish Psen2-coding sequence (represented as a protein sequence). Black arrows indicate the approximate positions of out-of-frame possible alternative start codons. No proteins produced by translation from such out-of-frame start codons would be detectable with the anti-GFP antibody used in B. The S4 residue affected by the S4Ter mutation is shown in red. The “acidic cluster” of residues at the N-terminal of zebrafish Psen2 as defined in Fig 3A of Sannerud et al. [52] is indicated by the blue background and the residues shaded grey in the protein alignment in that figure are indicated here by orange halos. The zebrafish fusion proteins are shown alongside a representation of human PSEN2 protein modified from the interactive diagram at Alzforum.org (version 2.4–2019, used with permission of FBRI LLC). Amino acid residues altered by known coding variants are color-coded as pathogenic (dark orange), non-pathogenic (green) or of uncertain pathogenicity (blue). Residue numbers, numbered transmembrane domains (TM), and the boundaries of numbered exons (Ex) containing the coding sequence are shown.
|