|
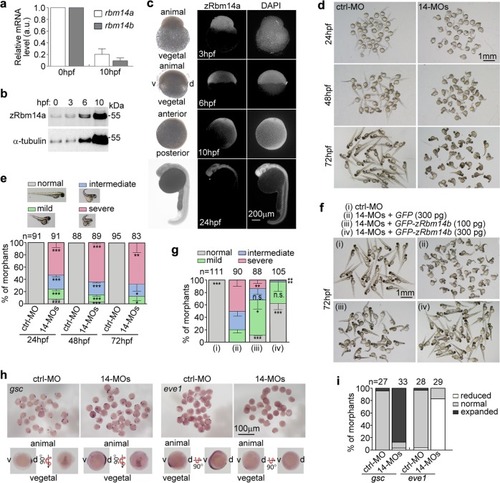
Zebrafish <italic>rbm14</italic> morphants display dorsalized phenotypes.arbm14 was downregulated during embryogenesis. Total mRNAs were extracted from 50 zebrafish embryos at 0 or 10 hpf. The expression levels of rbm14a and rbm14b were analyzed by qPCR. β-Actin was used as internal control. qPCR results from two independent experiments are presented as mean ± SD. b Total protein levels of zRbm14a increased during early embryonic development. Zebrafish embryos at the indicated stages were removed of the yolk and subjected to immunoblotting. Proteins from 8 embryos were loaded in each lane. c Immunofluorescent staining of zRbm14 in zebrafish embryos. Nuclear DNA was stained by DAPI. The animal and vegetal poles and ventral (v) and dorsal (d) sides are marked in the bright-field images. d Embryo morphologies at the indicated developmental stages. Zebrafish embryos at the one-cell stage were injected with a control morpholino oligonucleotide (ctrl-MO; 8 ng per embryo) or two MOs specific to rbm14a and rbm14b, respectively (14-MOs; 4 ng each per embryo) (see Supplementary Fig. 2a–e). e Quantification results for the experiments in d, based on the criteria and examples shown. f, g Exogenous zRbm14b rescued the dorsalized phenotypes. Zebrafish embryos at the one-cell stage were co-injected with the indicated MOs (total 8 ng per embryo) and in vitro-transcribed mRNA (also see Supplementary Fig. 1f). The morphants were examined at 72 hpf. Those injected with ctrl-MO served as negative control. Data in e and g, presented as mean ± SD, were from three independent experiments. Student’s t-test against the ctrl-MO-injected populations: n.s., no significance (P > 0.05); *P < 0.05; **P < 0.01; ***P < 0.001. Total numbers of embryos analyzed are listed over histograms. h, i The rbm14 morphants show defective dorsoventral patterning. Zebrafish embryos microinjected as in d were subjected to mRNA in situ hybridization at the 75–90% epiboly stages (h) and quantified (i). Total numbers of embryos analyzed are listed over each histogram
|