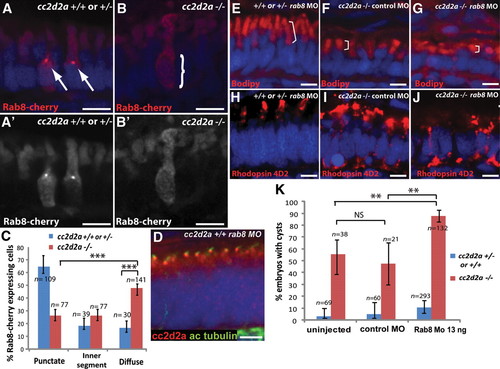
cc2d2a is required for Rab8 localization and interacts genetically with rab8 in ciliated cells. (A?B′) Expression of a transducin-promoter-driven Rab8-cherry construct in cc2d2a+/+ photoreceptors (A and A′) is concentrated in one punctum at the base of the outer segment (arrows), whereas it is diffuse in cc2d2a-/- photoreceptors (B and B′, bracket). (C) Proportion of Rab8-cherry expressing photoreceptors with punctate versus slightly enhanced inner segment or diffuse cellular expression (***P < 0.0001, χ2 test, bars indicate 95% confidence interval). (D) Cc2d2a localization (red) in 4 d.p.f. Rab8 morphant (rab8MO) retinas co-stained with acetylated tubulin to mark cilia. (E?J) Synergistic effect of partial rab8 knockdown on the retinal phenotype of cc2d2a-/- embryos. (E?G) Length of outer segments highlighted with bodipy counterstain (red, brackets) is reduced in rab8-morpholino-injected cc2d2a mutant embryos (G) compared with control-morpholino-injected mutants (F). At the same dose of rab8 morpholino, cc2d2a+/+ embryos have well-formed outer segments of normal length (E). (H?J) Rhodopsin localization (4D2 antibody, red) is virtually absent from remnant outer segments in rab8-morpholino-injected cc2d2a mutants (J) compared with control-morpholino-injected mutants (I). At the same dose of rab8 morpholino, there is no mislocalization of rhodopsin in cc2d2a+/+ siblings (H). (K) Histogram showing the proportion of embryos with pronephric cysts (red bars indicate cc2d2a-/- mutants and blue bars indicate wild-type and heterozygote siblings). Bars indicate 95% confidence intervals. Pairwise comparisons between rab8-morpholino-injected cc2d2a-/- fish and uninjected siblings, as well as rab8-morpholino-injected cc2d2a-/- and control (gbx2) morpholino-injected cc2d2a-/- fish are significant (**P = 0.0001, χ2 test). NS, non-significant. All images are cryosections of 5 d.p.f. retinas except (D), which is a cryosection of a 4 d.p.f. retina. Scale bars are 4 Ám in all panels. +/+ or +/- in (E) and (H) refers to cc2d2a+/+ or +/-.
|

