Fig. 1
- ID
- ZDB-FIG-110121-29
- Publication
- Yu et al., 2011 - The Cell Adhesion-associated Protein Git2 Regulates Morphogenetic Movements during Zebrafish Embryonic Development
- Other Figures
- All Figure Page
- Back to All Figure Page
|
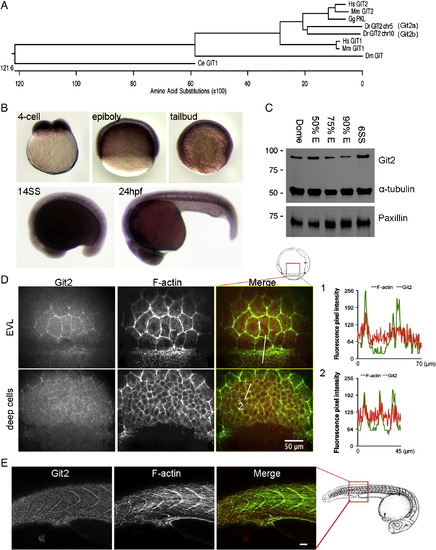
Identification and characterization of git2 genes in zebrafish. (A) Phylogenetic analysis of zebrafish git2 family genes. Dendogram of zebrafish git2a on chromosome 5 and git2b on chromosome 10 and related orthologs from other species. (B) In situ hybridization of git2a mRNA expression in the zebrafish embryo. git2a expression was ubiquitously detected at the 4-cell, epiboly, tailbud and 14-somite (14SS) and 24 hpf stages. (C) Western blotting of zebrafish Git2 protein at dome, 50%, 75%, 90% epiboly and 6-somite (6SS) stages, α-tubulin and paxillin were used as loading controls. (D) Immunohistochemistry of Git2 (red) at the 75% epiboly stage. Embryos were co-stained with phalloidin to detect F-actin (green). Images show surface EVL cells and deep cells (30 μm below the surface). Scale bar, 50 μm. Fluorescent intensity profiles show relative F-actin (green) and Git2 (red) levels in EVL cells at the blastoderm margin (1) and deep cells (2). (E) Immunohistochemistry of Git2 (red) at the 24 hpf. Embryos were co-stained with phalloidin to detect F-actin (green). Scale bar, 50 μm. Zebrafish embryo drawings are adapted from Kimmel et al., 1995. |
| Gene: | |
|---|---|
| Antibody: | |
| Fish: | |
| Anatomical Terms: | |
| Stage Range: | 4-cell to Prim-5 |
Reprinted from Developmental Biology, 349(2), Yu, J.A., Foley, F.C., Amack, J.D., and Turner, C.E., The Cell Adhesion-associated Protein Git2 Regulates Morphogenetic Movements during Zebrafish Embryonic Development, 225-237, Copyright (2011) with permission from Elsevier. Full text @ Dev. Biol.