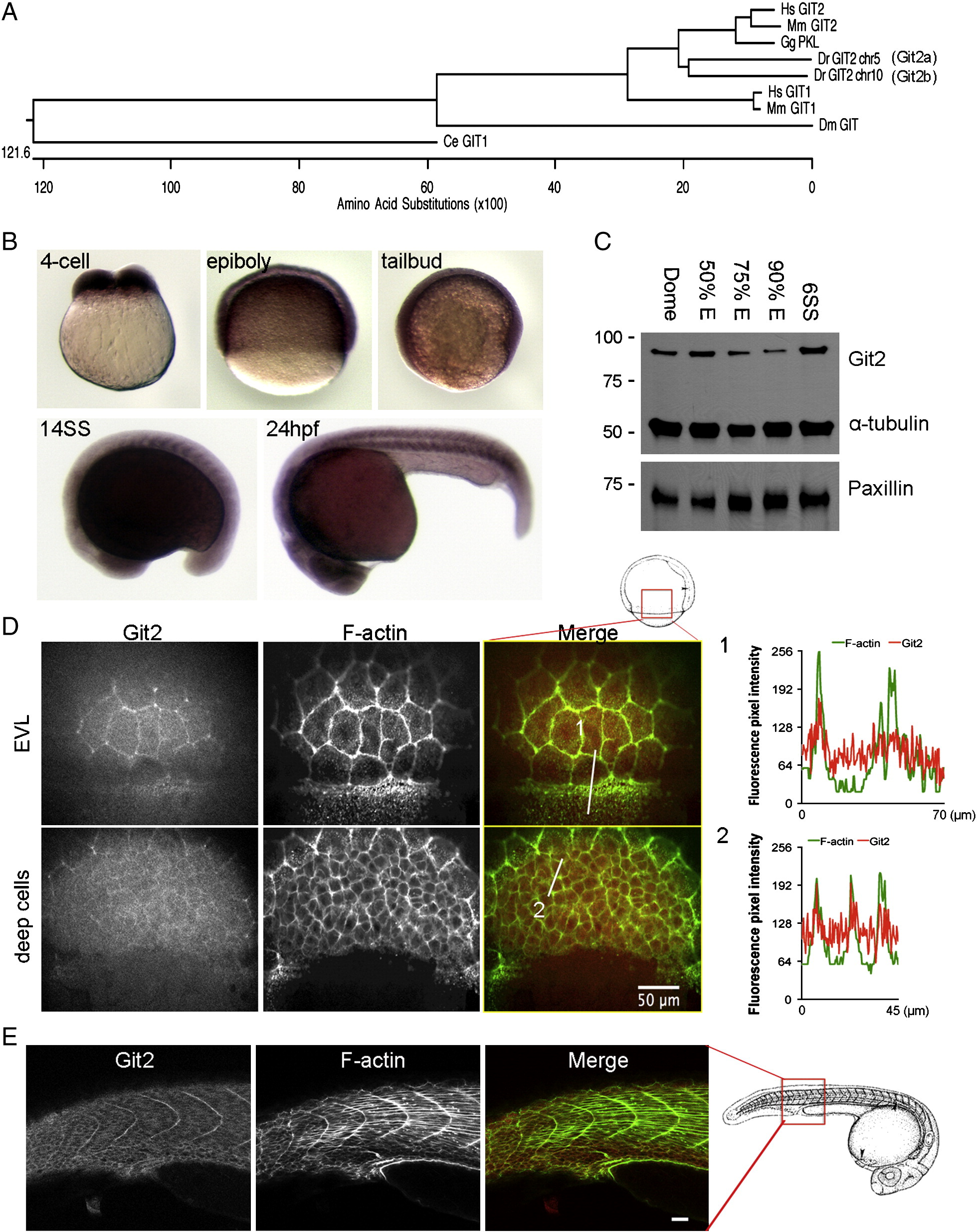
Fig. 1 Identification and characterization of git2 genes in zebrafish. (A) Phylogenetic analysis of zebrafish git2 family genes. Dendogram of zebrafish git2a on chromosome 5 and git2b on chromosome 10 and related orthologs from other species. (B) In situ hybridization of git2a mRNA expression in the zebrafish embryo. git2a expression was ubiquitously detected at the 4-cell, epiboly, tailbud and 14-somite (14SS) and 24 hpf stages. (C) Western blotting of zebrafish Git2 protein at dome, 50%, 75%, 90% epiboly and 6-somite (6SS) stages, α-tubulin and paxillin were used as loading controls. (D) Immunohistochemistry of Git2 (red) at the 75% epiboly stage. Embryos were co-stained with phalloidin to detect F-actin (green). Images show surface EVL cells and deep cells (30 μm below the surface). Scale bar, 50 μm. Fluorescent intensity profiles show relative F-actin (green) and Git2 (red) levels in EVL cells at the blastoderm margin (1) and deep cells (2). (E) Immunohistochemistry of Git2 (red) at the 24 hpf. Embryos were co-stained with phalloidin to detect F-actin (green). Scale bar, 50 μm. Zebrafish embryo drawings are adapted from Kimmel et al., 1995.
Reprinted from Developmental Biology, 349(2), Yu, J.A., Foley, F.C., Amack, J.D., and Turner, C.E., The Cell Adhesion-associated Protein Git2 Regulates Morphogenetic Movements during Zebrafish Embryonic Development, 225-237, Copyright (2011) with permission from Elsevier. Full text @ Dev. Biol.