Fig. 5
- ID
- ZDB-FIG-090804-59
- Publication
- Ng et al., 2009 - Zebrafish mutations in gart and paics identify crucial roles for de novo purine synthesis in vertebrate pigmentation and ocular development
- Other Figures
- All Figure Page
- Back to All Figure Page
|
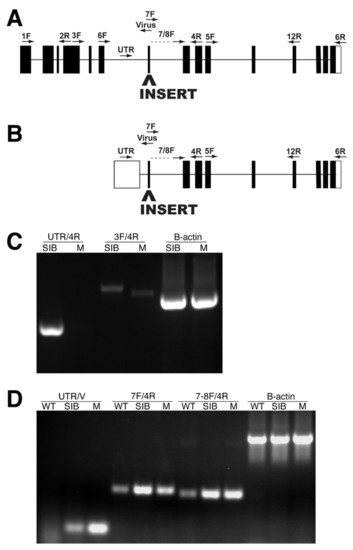
Molecular analysis of the paicshi2688 locus. (A,B) The approximate position of the proviral insertion and RT-PCR primers mapped onto a schematic of the mutated gene (www.ensembl.org/Danio_rerio/). (A) paicsLONG is encoded by at least 15 exons, and all of the conserved enzymatic domains of the Paics protein are found in the sequence encoded by exons 7-15. The proviral insert in paics mutants is located in exon 7 of paicsLONG. (B) paicsSHORT is composed of the latter nine exons of paicsLONG, in addition to a tenth noncoding first exon. The proviral insert in paics mutants is located in exon 2 of paicsSHORT. (C) RT-PCR using primer sets that amplify the region between exons 4 and 9 of paicsLONG (3F/4R), and exons 1 and 4 of paicsSHORT (UTR/4R) indicates that both transcripts are expressed in wild-type embryos. Amplification of the region surrounding the site of proviral insertion (3F/4R) in paicsLONG indicates that the proviral insert leads to an alteration in mRNA splicing in the paicsLONG transcript. (D) RT-PCR using primer sets that assay for the presence of the proviral insert in paicsSHORT (UTR/V) indicates that the viral insert is present in paicsSHORT transcripts. RT-PCR amplification using paicsSHORT-specific exon 2 primers located downstream of the site of proviral insertion (7F/4R and 7-8F/4R) indicates that the virus sequence in paicsSHORT does not lead to detectable changes in transcript levels. WT, wild-type AB; SIB, wild-type sibling mixture (+/+ and +/-); M, homozygous mutant. |