- Title
-
A single base pair substitution in zebrafish distinguishes between innate and acute startle behavior regulation
- Authors
- Ortiz, E.A., Campbell, P.D., Nelson, J.C., Granato, M.
- Source
- Full text @ PLoS One
|
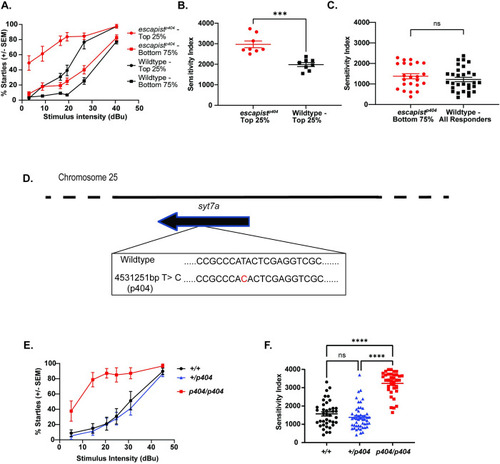
RNA sequencing linkage analysis identifies a single base pair change on Chromosome 25 that is tightly linked to the (A) Acoustic sensitivity curve for |
|
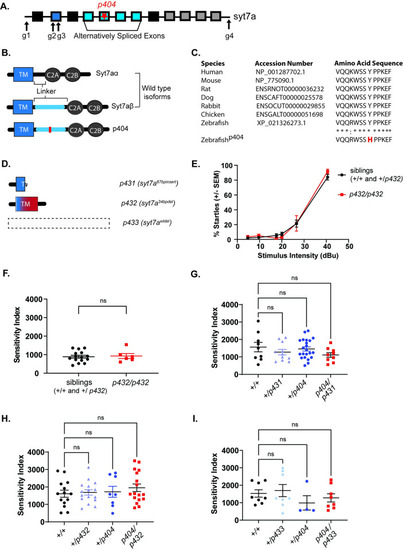
CRISPR-Cas9 generated (A) Schematic of |
|
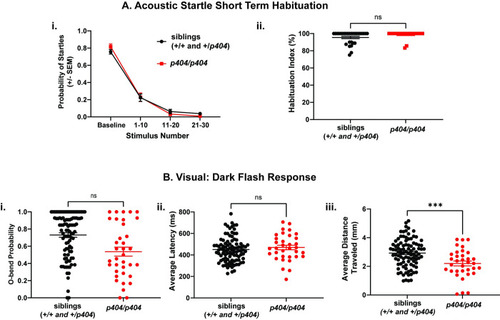
(A) Acoustic startle short term habituation assay. i) Percent startle of both siblings (n = 20 larvae, black) and |
|
(A) Acoustic startle short term habituation assay: i) Average probability of startle for siblings (n = 29 larvae, black) and |