- Title
-
Genetic and chemical disruption of amyloid precursor protein processing impairs zebrafish sleep maintenance
- Authors
- Özcan, G.G., Lim, S., Canning, T., Tirathdas, L., Donnelly, J., Kundu, T., Rihel, J.
- Source
- Full text @ iScience
|
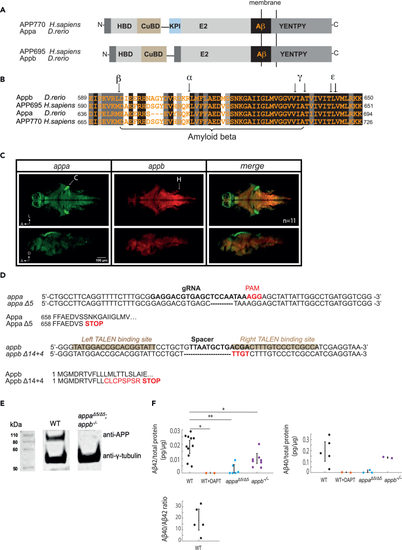
Zebrafish App protein organization, gene expression, and mutant generation (A) There are 2 (B) Alignment of Aβ regions of zebrafish Appa and Appb to human APP695 and APP770 shows high conservation within the Aβ region and the proteolytic cleavage sites (indicated with black arrows): α-secretase cleavage site (α), β-secretase cleavage site (β), γ-secretase cleavage sites (γ), and ε-cleavage sites (ε). Black, dark gray, and light gray boxes indicate strictly, highly, and moderately conserved amino acid residues, respectively. (C) As detected by multiplexed hybridization chain reaction (HCR), (D) CRISPR/Cas9 targeting of zebrafish (E) Western blot analysis of APP in brain homogenates from wild-type (WT) and (F) Elisa detection of Aβ42 (left) and Aβ40 (right) levels in adult brain homogenates from WT controls, WT animals treated with the γ-secretase inhibitor DAPT for 24 h, |
|
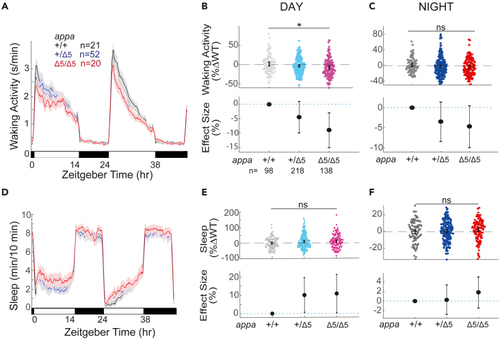
Exemplar 48 h traces of average waking activity taken from a single experiment of (B) Day waking activity and C) night waking activity for (D) Exemplar 48 h traces of average sleep for the same experiment shown in (A). (E) Day sleep and (F) night sleep of WT, heterozygous, and |
|
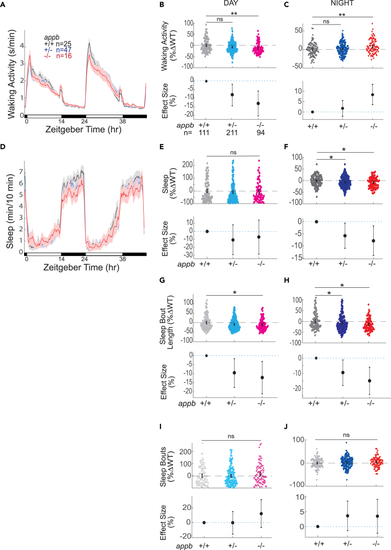
Exemplar 48 h traces of average waking activity taken from a single experiment of (B) Day waking activity and (C) night waking activity for (D) Exemplar 48 h traces of average sleep for the same experiment shown in (A). (E) Day sleep, (F) night sleep, (G) day sleep length, (H) night sleep length, (I) day sleep bout number, and (J) night sleep bout number of WT |
|
The γ-secretase inhibitor DAPT shortens sleep bout lengths at night in WT but not in (A and B) Exemplar 48 h traces on a 14h:10h light:dark cycle of the average waking activity of WT (A) and (C and D) Exemplar 48 h traces of the average sleep from the same experiment shown in (A) and (B). (E) Night sleep and (F) night sleep bout length of WTs and |
|
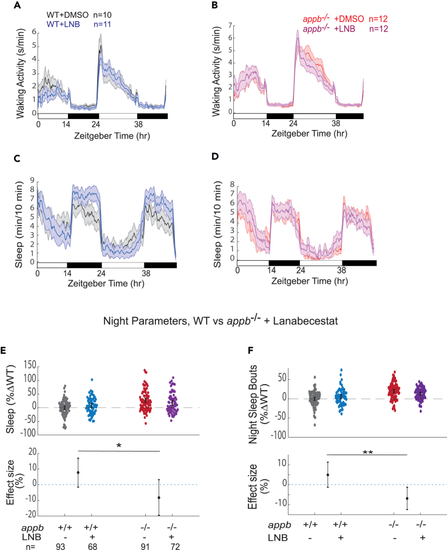
The β-secretase inhibitor lanabecestat increases sleep at night in WT but not in (A and B) Exemplar 48 h traces on a 14h:10h light:dark cycle of the average waking activity of WT (A) and appb−/− mutants (B) continuously exposed to either 0.3 μM lanabecestat or DMSO vehicle control. (C and D) Exemplar 48 h traces of the average sleep from the same experiment shown in A and B. (E) Night sleep and (F) night sleep bout length of WTs and |
|
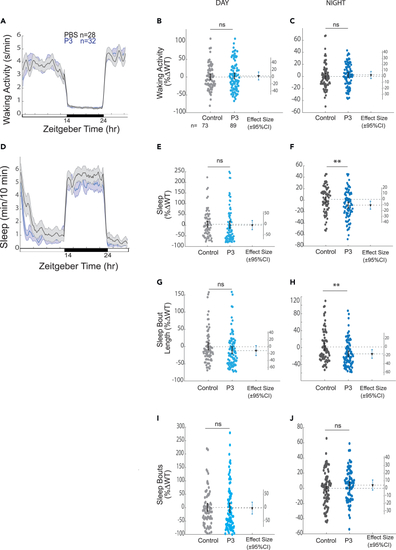
Intraventricular injection of P3 decreases sleep and shortens sleep bout lengths Exemplar 24 h traces of the average waking activity of WT larvae injected with either P3 or vehicle control on a 14h:10h light:dark cycle. Shown is a single exemplar experiment. (B and C) Average waking activity during the day (B) and night (C) across three independent experiments. Each dot represents a single larva normalized to the experiment-matched WT mean. The bars represent the mean. (D) Exemplar 24 h trace of the average sleep for the experiment shown in (A). (E) Day sleep, (F) night sleep, (G) day sleep bout length, (H) night sleep bout length, (I) day sleep bout number, and (J) night sleep bout number. n = the number of larvae. nsp > 0.05, ∗p ≤ 0.05, ∗∗p ≤ 0.01, Kruskal-Wallis, Tukey’s post hoc test. n = number of larvae. |