- Title
-
The synaptic receptor Lrp4 promotes peripheral nerve regeneration
- Authors
- Gribble, K.D., Walker, L.J., Saint-Amant, L., Kuwada, J.Y., Granato, M.
- Source
- Full text @ Nat. Commun.
|
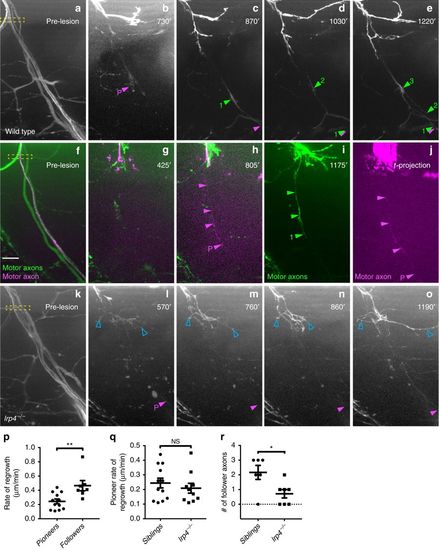
Pioneering axons establish regenerative path for follower axons. a?e Images from time lapse movie showing early regeneration of wild-type motor nerve with all axons expressing GFP. a Pre-lesion image; yellow box marks transection site. b A pioneering axon (magenta arrowhead and ?P?) crosses the injury gap ~800?min post-injury and extends ventrally. c, d, e Follower axons fasciculate with pioneering axon (numbered green arrowheads). f?j Still images from time lapse movie showing early regeneration of 5 dpf wild-type motor nerve; all axons express GFP, single-axon expresses mKate (magenta). f Pre-lesion image; yellow box marks transection site. Scale bar is 10?Ám. g Axon sprouts in proximal nerve stump start extending and retracting ~400?min post transection. h Single-pioneering axon (magenta) crosses the injury gap and extends ventrally along distal nerve by 800?min post transection. Magenta arrowheads mark pioneer axon, which grows at a rate of 0.27?Ám/min. i Follower axon extends and grows along pioneering axon; green arrowheads mark follower path, which grows at a rate of 0.36?Ám/min. j By the end of imaging, the magenta pioneer axon has extended ventrally, as shown via a maximum projection image across time (magenta arrowheads). k?o Still images from time lapse movie showing early regeneration of lrp4 mutant motor nerve labeled with GFP. k Pre-lesion image; yellow box marks transection site. l Early growth cones sprout from proximal stump (open blue arrowheads) and pioneer axon extends ventrally ~600?min post transection (magenta arrowhead and ?P?). m?o Throughout imaging, only the pioneer axon extends to ventral myotome; follower axons explore transection site but do not traverse injury gap (open blue arrowheads). p Quantification of pioneer axon growth rate and follower axon growth rate in wild-type siblings. Pioneer axon average rate of regrowth is 0.24?Ám/min (n?=?12 nerves in 8 larvae); follower axon average rate of regrowth is 0.47?Ám/min (n?=?7 nerves in 4 larvae; unpaired t-test p?=?0.0055; t?=?3.181, df?=?17; error bars show mean and SEM). q Quantification of pioneer axon rate of regrowth in siblings and lrp4 mutants. Pioneer axons grow at equivalent rates: sibling average rate of regrowth?=?0.24?Ám/min (n?=?12 nerves in 8 larvae); lrp4?/? average rate of regrowth?=?0.21?Ám/min (n?=?10 nerves in 4 larvae; unpaired two-tailed t-test p?=?0.4702, t?=?0.7361, df?=?20; error bars show mean and SEM). r Number of follower axons in siblings and lrp4 mutants. In siblings, multiple follower axons extend (average number of follower axons?=?2.2, n?=?6 nerves in 2 larvae) while in lrp4 mutants, follower axons fail to extend ventrally (lrp4 mutant average number of follower axons?=?0.7, n?=?7 nerves in 2 larvae). Number of follower axons in siblings versus mutants is significantly different (two-tailed t-test p?=?0.0206, t?=?2.703, df?=?11; error bars show mean and SEM) PHENOTYPE:
|
|
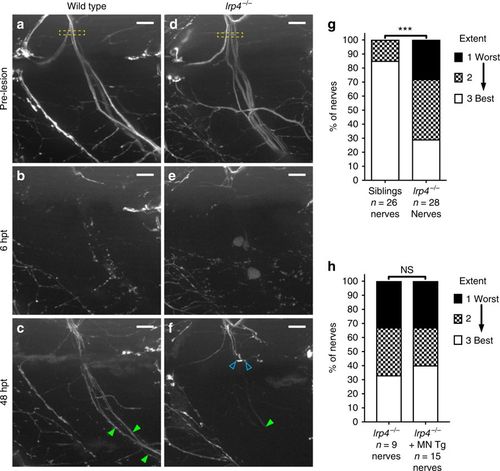
lrp4 is required non-cell-autonomously for axonal growth during regeneration. a Lateral view of a wild-type motor nerve labeled with GFP before laser transection at 5 dpf, with transection site marked by yellow box. Scale bar is 10?Ám. b Six hours post transection (hpt), motor axons distal to the transection site have fully degenerated. c Forty-eight hours post transection, motor axons have regrown along their original path; green arrowheads indicate multiple regenerated fascicles; this representative nerve received a score of ?best? regeneration. d Lateral view of lrp4 mutant motor nerve labeled with GFP before laser transection at 5 dpf, with transection site marked by yellow box. e Six hours post transection, distal motor axons have fully degenerated. f Forty-eight hours post transection, motor axons have sprouted from the proximal stump but most fail to cross the injury site (open blue arrowheads); green arrowhead indicates stalled fascicle; this representative nerve received a score of ?worst? regeneration. g Quantification of nerve regeneration in wild type and lrp4 mutant larvae. In wild type, 85% of nerves regenerate ?best? with two or more distinct fascicles (n?=?26 nerves from 13 larvae). In lrp4 mutants, 70% of nerves regenerate zero or one fascicle (n?=?28 nerves from 13 larvae) by 48?h post transection. Wild-type siblings regenerate significantly better than lrp4 mutants (?2-test p?<?0.0001, ?2?=?18.48, df?=?2). h Quantification of nerve regeneration in lrp4 mutant larvae and lrp4 mutant larvae expressing the motor neuron-specific mnx1:Lrp4-GFP transgene. At 48 hpt, lrp4 mutant larvae expressing Tg(mnx1:Lrp4-GFP) regenerate poorly (n?=?15 nerves from 6 larvae), like lrp4 mutant larvae (n?=?9 nerves from 4 larvae; ?2-test p?=?0.9266, ?2?=?0.1524, df?=?2) |
|
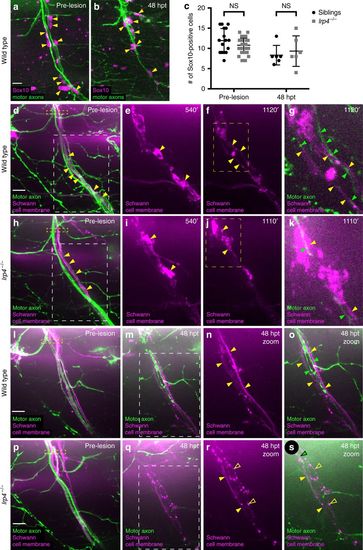
Schwann cell morphology during early regeneration is disrupted in lrp4 mutants. a Pre-transection 5 dpf, GFP-labeled sibling nerve stained with anti-Sox10 antibody labeling Schwann cell nuclei (magenta). Yellow arrowheads mark Schwann cell nuclei; scale bar 10?Ám. b Sibling nerve 48 hpt labeled with GFP and stained with anti-Sox10 antibody. Yellow arrowheads mark Schwann cell nuclei. c Quantification of Schwann cell nuclei along individual nerves pre-transection and 48 hpt in siblings and lrp4 mutants. Pre-transection average Schwann cell number in siblings?=?11.9 per nerve, n?=?16 nerves; average Schwann cell number in lrp4 mutants?=?10.78 per nerve, n?=?36 nerves (two-tailed t-test p?=?0.08, t?=?1.743, df?=?50; error bars denote mean and SD). Average Schwann cell number 48 hpt in siblings?=?8.3 per nerve, n?=?6 nerves; average Schwann cell number 48 hpt in lrp4 mutants?=?9.3 per nerve, n?=?6 nerves (two-tailed t-test p?=?0.59, t?=?0.5459, df?=?10; error bars denote mean and SD). d?k Images from time lapse movies showing early regeneration dynamics in sibling (d?g) and lrp4 mutant (h?k) nerves. Motor axons express GFP, Schwann cells express mRFP (magenta). d Pre-transection sibling nerve, yellow box marks transection site. Schwann cell membranes (yellow arrowheads) are thin and closely associate with motor axons. White box indicates region magnified in (e?f). Scale bar 10?Ám. e Magnified image showing granular Schwann cell morphology characteristic of a denervated Schwann cell (yellow arrowheads). f Schwann cell membranes revert to smooth pre-injury morphology (yellow arrowheads). Yellow box marks region magnified in (g). g Magnified image at same timepoint as (f), showing regenerating axons (green arrowheads) associating with flattened Schwann cell membranes (yellow arrowheads; n?=?4/4 nerves). h Pre-transection lrp4 mutant nerve, yellow box marks transection site. Schwann cell membranes (yellow arrowheads) look wild type. White box indicates region magnified in (i?j). i Magnified image showing granular Schwann cell morphology (yellow arrowheads). j Lrp4 mutant Schwann cell membranes fail to revert to smooth pre-injury morphology (yellow arrowheads). Yellow box marks region magnified in (k). k Magnified image at same timepoint as j, showing a regenerating axon (green arrowhead) associating with granular Schwann cell membranes (yellow arrowheads; n?=?7/8 nerves). l Pre-transection sibling nerve; yellow box marks transection site. m?o By 48 hpt, Schwann cell membranes form two thin, continuous tracks along which regenerating axons grow; white box indicates region magnified in n?o (filled yellow arrowheads mark Schwann cell membranes, green arrowheads mark regenerating axons; n?=?23/24 nerves). p Pre-transection lrp4 mutant nerve; yellow box marks transection site. q?s By 48 hpt, Schwann cell membranes fail to form two thin, continuous tracks and remain granular; white box in (q) indicates the region magnified in r?s (filled yellow arrowheads mark continuous Schwann cell membrane track; open yellow arrowheads mark granular membranes; green arrowhead marks regenerating axon; n?=?18/24 nerves) |
|
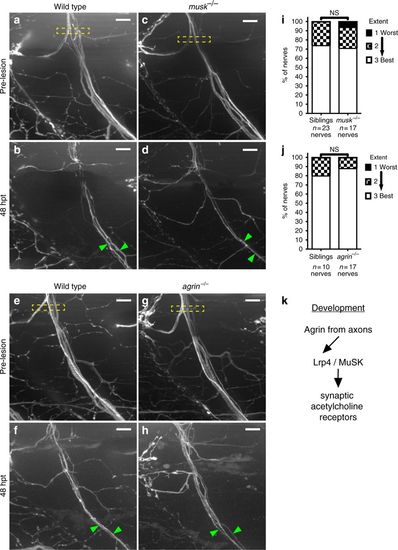
lrp4 acts in a MuSK-independent pathway to promote axonal regeneration. a Lateral view of wild-type GFP-labeled motor nerve before laser transection at 5 dpf. Yellow dashed box indicates transection site, scale bar is 10?Ám. b Forty-eight hours post transection, multiple axon fascicles have regenerated ventrally. Green arrowheads indicate regenerated fascicles. c Lateral view of musk mutant motor nerve before laser transection at 5 dpf. Yellow dashed box indicates transection site. d Forty-eight hours post transection, motor axons have regenerated ventrally. Green arrowheads indicate regenerated fascicles. e Lateral view of wild-type GFP-labeled motor nerve before transection. f Forty-eight hours post transection, multiple axon fascicles regenerate ventrally. Green arrowheads mark regenerated fascicles. g Lateral view of agrin mutant motor nerve before transection at 5 dpf. Yellow box indicates transection site. h Forty-eight hours post transection, agrin mutant motor axons have regenerated ventrally. Green arrowheads mark regenerated fascicles. i Quantification of nerve regeneration in wild-type siblings (n?=?23 nerves from 11 larvae) and musk mutants (n?=?17 nerves from 11 larvae) at 48?h post transection. musk mutant motor nerves regenerate as well as wild-type sibling motor nerves (Fisher?s exact test p?=?0.4982). j Quantification of nerve regeneration in wild-type siblings (n?=?10 nerves from 4 larvae) and agrin mutants (n?=?17 nerves from 6 larvae). Agrin mutant motor nerves regenerate as well as wild-type sibling nerves (Fisher?s exact test p?=?0.6125). k Schematic of the canonical Agrin-Lrp4-MuSK pathway critical for neuromuscular synapse formation |
|
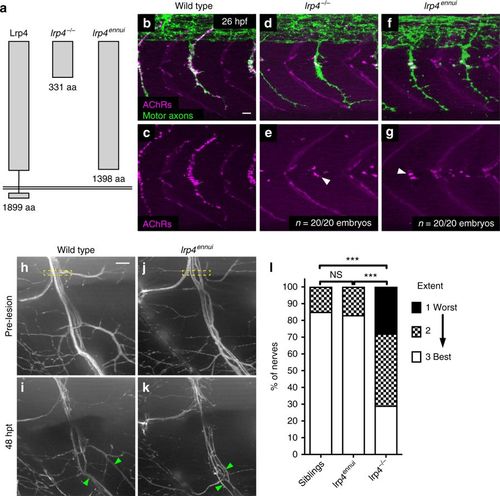
A truncated version of Lrp4 promotes nerve regeneration. a Schematic of lrp4 alleles and their protein structures: wild-type full-length Lrp4 is shown at far left (1899 amino acids). lrp4?/? allele encodes a protein of 331 amino acids. lrp4ennui encodes a protein of 1398 amino acids that is predicted to contain almost the entire ectodomain but lack the transmembrane and intracellular domains. b?g Lateral views of wild type, lrp4?/?, and lrp4ennui embryos at the stage when en passant neuromuscular synapses form (26 hpf) stained for acetylcholine receptors (AChRs; magenta) and motor axons (green). Scale bar is 10?Ám. b?c In wild-type embryos, AChR clusters assemble beneath the entire motor axon. d?g In lrp4 mutant and lrp4ennui embryos there is a strong reduction in AChR clustering beneath the motor axon, with only a few AChR clusters observed (white arrowheads); n?=?20 out of 20 embryos examined for each genotype. h Lateral view of pre-transection wild-type motor nerve labeled with GFP at 5 dpf. Transection site is marked by yellow box. Scale bar is 10?Ám. i Forty-eight hours post transection in wild-type larvae, multiple fascicles have regenerated ventrally (marked by green arrowheads). j Lateral view of pre-transection motor nerve in lrp4ennui labeled with GFP at 5 dpf. Transection site marked by yellow box. k Forty-eight hours post transection in lrp4ennui, multiple fascicles have regenerated ventrally (green arrowheads). l Quantification of nerve regeneration in wild-type larvae (n?=?26 nerves from 13 larvae), lrp4 mutants (n?=?28 nerves from 13 larvae) and lrp4ennui (n?=?24 nerves from 9 larvae) at 48?h post transection. lrp4ennui nerves regenerate to the same extent as wild-type sibling nerves (wild-type siblings ?best? regeneration?=?85% of nerves; lrp4ennui ?best? regeneration?=?83% of nerves; ?2-test p?=?0.1672, ?2?=?3.577, df?=?2) |
|
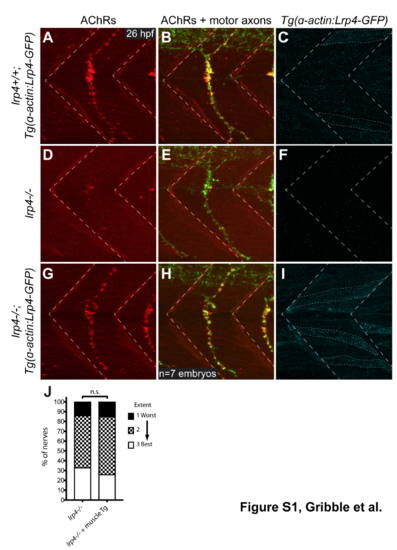
Selective expression of lrp4 in muscle cells rescues neuromuscular synaptic defects in lrp4 mutants (A) Lateral view of single hemisegment in a 26 hpf wild-type zebrafish embryo expressing Tg(?-actin:Lrp4-GFP), showing normal distribution of acetylcholine receptors (AChRs) in the center of the hemisegment. (B) Merged image of AChRs and motor axons labeled with znp-1 antibody, showing AChRs clustered beneath motor axon terminals. (C) Localization and expression of Tg(?-actin:Lrp4-GFP) in wild-type muscle cells. Lrp4-GFP localizes in puncta distributed throughout each muscle cell. (D,E) lrp4 mutants at 26 hpf show a dramatic reduction in AChR clusters and neuromuscular synapses, but motor axons extend normally. (F) No Lrp4-GFP signal is observed in this mutant, and the muscle rescue transgene is not detectable by PCR. (G,H) lrp4 mutants expressing Tg(?- actin:Lrp4-GFP) show AChR clusters and neuromuscular synapses at 26 hpf that appear wild-type (n=7 embryos). (I) Expression of Tg(?-actin:Lrp4-GFP) in muscle cells of lrp4 mutant embryo. (J) Quantification of nerve regeneration in lrp4 mutant larvae and lrp4 mutant larvae expressing the muscle-specific ?-actin:Lrp4-GFP transgene. At 48 hpt, lrp4 mutant larvae expressing Tg(?-actin:Lrp4-GFP) regenerate poorly (n=27 nerves), like lrp4 mutant larvae (n=21 nerves; Chi-square test p=0.8505; Chi-square=0.3238, df=2). |
|
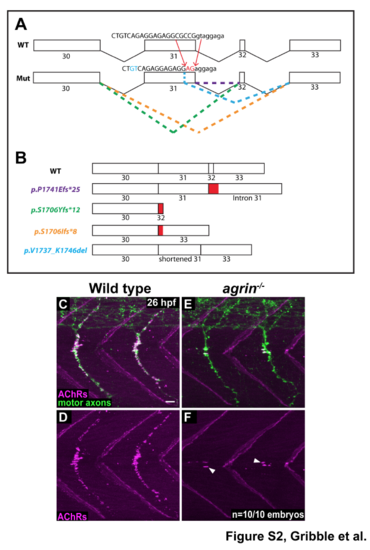
Zebrafish agrin is required for neuromuscular synapse formation (A) Summary of transcripts generated through CRISPR/Cas9 injection. The allele used for all experiments was a 7bpDEL;2bpINS at the end of exon 31, which resulted in loss of the exon 31 splice donor site. Wild type sequence is flanked by red arrows, and the genomic loss is marked in the mutant sequence just below. Sequencing cDNA from pools of mutants and siblings yielded four predominant transcripts, whose alternative splicing is shown using green, orange, blue, and purple dashed lines. (B) Exon structure and resulting protein sequence for wild type agrin and the four predominant transcripts identified from sequencing cDNA. Three of four transcripts result in a frameshift and premature stop codon (purple, green, and orange transcripts), while one transcript is an in-frame deletion (blue transcript). All four transcripts result in severe swimming defects in zebrafish larvae at 36 hpf (data not shown). (C-D) At 26 hpf in wild type embryos, neuromuscular synapses (AChR labeling in magenta) are abundant beneath motor axons (green). (E-F) In agrin mutants, neuromuscular synapses are significantly reduced; white arrowheads show residual AChR clusters on the muscle cells at the horizontal myoseptum. N = 10 out of 10 embryos analyzed. |

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. |