- Title
-
Hematopoietic Stem Cell Arrival Triggers Dynamic Remodeling of the Perivascular Niche
- Authors
- Tamplin, O.J., Durand, E.M., Carr, L.A., Childs, S.J., Hagedorn, E.J., Li, P., Yzaguirre, A.D., Speck, N.A., Zon, L.I.
- Source
- Full text @ Cell
|
A Transgenic Zebrafish Line that Specifically Marks HSPCs (A) Single frame of time-lapse (hr post fertilization:min) showing Runx:mCherry+ HSPCs (arrowheads, red nuclei) budding from kdrl:GFP+ hemogenic endothelium of DA (green). See Movie S1. (B?D) CHT is colonized by Runx:GFP+ HSPCs (green) that are closely associated with kdrl:mCherry+ ECs (red). Caudal artery (CA) is dorsal to CHT, caudal vein (CV) is ventral, and circulation runs posterior (right arrow) and anterior (left arrow), respectively. Cluster of three Runx:GFP+ high cells outlined with dashed line and four Runx:GFP+ low cells indicated with arrowheads. 58 hpf embryo. (E?G) Runx:GFP+ high and low cells quantified, with CHT length as indicator of stage (anterior from cloaca to posterior limit of CA). Note: confocal images are 3D rendered depth or max projections of 20?30 µm z stacks. Scale bars, (A) 15 µm; (B) 25 µm. Error bars show mean ± SEM. See also Figure S1. |
|
Endothelial Cells in the Perivascular Niche Remodel to Surround a Single HSPC (A) Four frames from time-lapse Movie S2 40?42 hpf (hr post fertilization:min). Upper row is a merge of Runx:GFP+ HSPC (green, middle row) and kdrl:RFP ECs (red, lower row). A group of surrounding ECs (broken circle) remodel around a single HSPC soon after its arrival. (B) Higher magnification (60×) live image of single Runx:GFP+ HSPC surrounded by kdrl:mCherry ECs at 78 hpf (orthogonal views). (C) 3D rendered projection of scanning confocal image from fixed 80 hpf embryo. (i) merge, (ii) kdrl:mCherry ECs, (iii) Runx:GFP+ HSPCs, (iv) DRAQ5 nuclei, and (v) kdrl:mCherry projection with 3D modeled green HSPCs and five blue surrounding EC nuclei (arrowhead indicates HSPC in EC surround). All views: dorsal up, ventral down. Scale bars, 10 µm. See also Figure S3. |
|
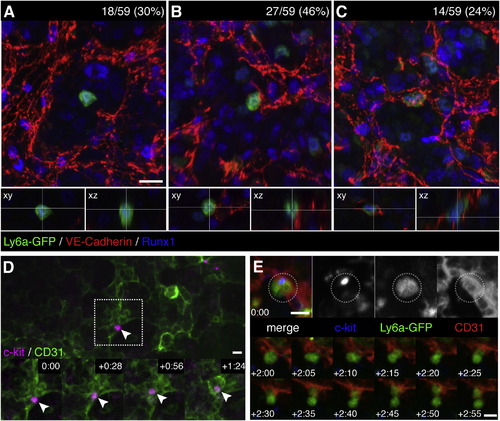
Endothelial Cells Surround HSPCs in the Fetal Liver Microenvironment (A?C) FLs from E11.5 Ly6a-GFP mice were fixed and stained for immunofluorescence with anti-VE-Cadherin (red), anti-Runx1 (blue), and anti-GFP (green) antibodies. We scored 59 Ly6a-GFP+/Runx1+ cells from 3 FLs and identified 3 different HSPC-EC configurations: (A) abluminal with no contact between HSPC and ECs (18/59; 30%); (B) EC contact on one side of the HSPC (27/59; 46%); and (C) HSPC surrounded on all sides with ECs (14/59; 24%). See Movie S3. (D) c-kit+ cell (magenta) adhered to CD31+ ECs (green) in one lobe of an E11.5 FL (arrowhead). White box marks details below. Time-lapse frames show in <90 min the HSPC migrates into a field of ECs. Soon after, ECs surround HSPC to form niche. See also Movie S4. (E) c-kit+(blue)/Ly6a-GFP+(green) HSPC adhered to abluminal side of CD31+ EC (red). Following this cell >2 hr (1 frame/5 min) shows a division with distal and proximal daughters relative to sinusoid; the latter remains in an endothelial surround. See also Movie S5. Confocal images: 3D rendered depth projection (A, B, C, and E), orthogonal view (A, B, and C below), maximum projection (D) of z stack. Scale bars, 10 µm. See also Figure S4. |
|
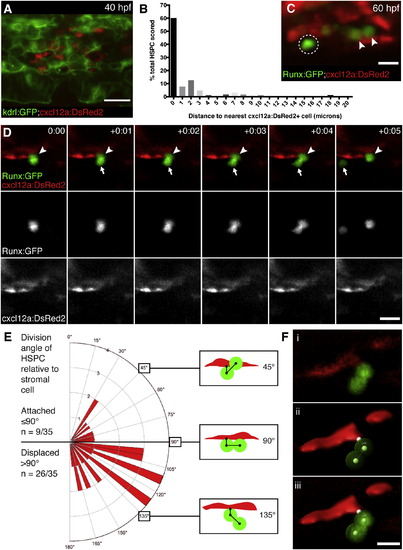
HSPCs Are Anchored to Perivascular Stromal Cells during Cell Divisions (A) cxcl12a:DsRed2+ stromal cells (red) underlie kdrl:GFP+ ECs (green). 40 hpf embryo. (B) Percentage of HSPCs in CHT scored by distance to nearest stromal cell (n = 168 total cells from 25 embryos). (C) Detail of Runx:GFP+ HSPCs (green) in proximity to cxcl12a:DsRed2+ stromal cells (red). Arrowheads mark HSPCs in contact with stromal cell. Circle marks HSPC with 2 µm gap between it and stromal cell. 60 hpf embryo. (D) Six frames selected from time-lapse Movie S6 (hr:min). Top row is a merge of Runx:GFP+ HSPC (green, middle row) and cxcl12a:DsRed2+ stromal cells (red, lower row). Proximal HSPC anchored to stromal cell (arrowhead) divides and releases distal daughter cell into circulation (arrow). (E) Rose diagram showing division plane of HSPC oriented relative to stromal cell. Majority of divisions result in displaced daughter cell (n = 26/35 cell divisions from 22 embryos; 95% confidence interval 0.567?0.875; mean division angle of 110°). Diagrams show HSPCs dividing over stromal cell surface (45°), perpendicular to stromal cell (90°), or displaced away from stromal cell (135°). Angles may be affected by release into flow of circulation. (F) 3D models used to measure HSPC divisions relative to stromal cells. (i) Volume rendered confocal image. (ii) 3D model showing angle measurement between attachment point on stromal cell surface and center points of proximal and distal HSPCs (example shown is 110°). (iii) Overlay of confocal image and 3D model. Scale bars, (A) 25 µm; (C and F) 10 µm; (D) 15 µm. |
|
High-Resolution Electron Microscopy of Endogenous HSPC in the Perivascular Niche (A) Last frame of CHT time lapse (60 hpf). Arrowhead marks HSPC lodged >6 hr. Runx:GFP (green), kdrl:mCherry (red), bright field (blue). Anterior left, posterior right, dorsal top, ventral bottom. (B) Detail of region in (A) marked by box. (C) Single section and orthogonal slice from serial block face EM scans. Lodged HSPC (purple, arrowhead), surrounding EC nuclei (green, numbered), and stromal cells (dark and light blue). (D) High-resolution EM of HSPC lodged in perivascular niche ventral to DA. The HSPC is in direct contact with one stromal cell (see higher-magnification inset). (E?J) Selected sections (left) through niche with cell membrane traces used to build 3D models (right). (E) In this section, the HSPC (purple) is mostly surrounded by EC (orange). Portions of fibroblastic (yellow) and stromal (light and dark blue) cells are visible. (F) About half of the HSPC surface is wrapped by EC. (G) The HSPC directly contacts the stromal cell. Portions of the fibroblastic cell, EC, and second stromal cell are visible. (H) Only the midsection of the HSPC contacts the stromal cell. (I) The fibroblastic cell surrounds the HSPC. Portions of two stromal cells are visible. (J) Most of the HSPC surface is wrapped by the fibroblastic cell. Scale bars, (B) 25 µm; (C) 10 µm; (D) 2 µm; and inset, 1 µm. See also Figure S5 and Movie S7. |
|
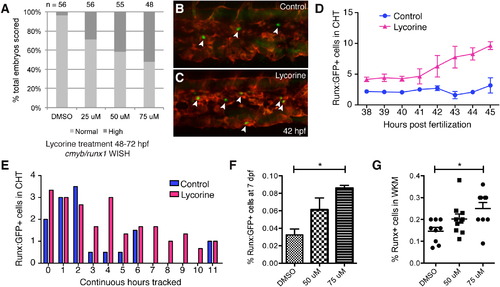
Modulating HSPC Niche Colonization with Lycorine Has Long-Term Effects on the Stem Cell Pool (A) Lycorine treatment dose-dependently increases the percentage of embryos with high cmyb/runx1 expression levels, as shown in Figure S6E (linear regression of Y [response] on X [dose] is significant: r2 = 0.146607, p = 6.46 × 109). (B and C) Stage-matched frames from parallel time-lapse movies show more HSPCs (arrowheads) with (C) 25 µM lycorine treatment than (B) DMSO-treated controls (42 hpf, Runx:GFP;kdrl:RFP). (D) HSPCs counted in each frame and then averaged each hour. Five movies imaged in parallel: n = 2 controls and n = 3 lycorine treated. (E) HSPCs in the same embryos were scored for continuous hours tracked in CHT. Longer median time HSPCs spent in CHT of lycorine-treated embryos was significant (1.67 hr; Wilcoxon signed-rank test, p = 0.01; HSPC counts normalized because treatment group was n = 3 and control group was n = 2). (F and G) Pools of Runx:GFP+ embryos treated with lycorine from 2?3 dpf and washed off had significantly increased HSPC at (F) 7 dpf (DMSO versus 75 µM, p = 0.0004; flow cytometry analysis of four independent pools per dose) and (G) 4 months (DMSO versus 75 µM, p = 0.006). Error bars show mean ± SEM. See also Figure S6. |
|
Characterization of Stable Runx:GFP and Runx:mCherry Transgenic Zebrafish Lines, Related to Figure 1 (A?F) Intercrosses of transgenic lines to show overlapping populations. Wide-field view of CHT at 72 hr postfertilization (hpf) with merged (i) GFP, mCherry and DIC channels, or (ii) GFP and mCherry. Detail in (ii) is shown below as (iii) merged GFP and mCherry, (iv) GFP, or (v) mCherry. Anterior left, posterior right, dorsal up, ventral down. Scale bars: (i-ii) 100 µm, (iii-v) 30 µm. (A) Overlap of Runx:GFP and Runx:mCherry lines. (B) Quantification of Runx:GFP and Runx:mCherry overlap. Runx:GFP overlaps 92 ± 11% with Runx:mCherry. Runx:mCherry overlaps 13 ± 6% with Runx:GFP. (CHT of n = 11 embryos scored). (C) Overlap of cd41:GFP and Runx:mCherry lines. (D) Quantification of cd41:GFP and Runx:mCherry overlap. cd41:GFP overlaps 60 ± 12% with Runx:mCherry. Runx:mCherry overlaps 44 ± 8% with cd41:GFP. (CHT of n = 12 embryos scored). cd41:gfp has known broader expression in thrombocyte progenitors at this stage (Bertrand et al., 2008 and Lin et al., 2005). (E) Overlap of cmyb:GFP and Runx:mCherry lines. (ii) Note the expression of the cmyb:GFP transgene in the ventral neural tube (arrow) and posterior notochord (arrowhead). (F) Quantification of cmyb:GFP and Runx:mCherry overlap. cmyb:GFP overlaps 18 ± 5% with Runx:mCherry. Runx:mCherry overlaps 35 ± 9% with cmyb:GFP. (CHT of n = 12 embryos scored). (G) Schematic of 60 hpf zebrafish embryo with location of thymus, dorsal aorta and caudal hematopoietic tissue (CHT). (H) Overlap of cd41:GFP and Runx:mCherry lines in the developing thymus. Note the bright circulating thrombocyte above. (I) Overlap of cmyb:GFP and Runx:mCherry lines in the developing thymus. (J) Forward scatter (FSC) and side scatter (SSC) were used to distinguish characteristic whole kidney marrow (WKM) populations (Traver et al., 2003). Runx:GFP+ cells were found predominately in the lymphoid/HSPC gate. (K) Flow cytometry of WKM from adult Runx:GFP transgenic zebrafish. Runx:GFP+ cells represent on average 0.133% of the total WKM. (L) Immunohistochemistry with anti-GFP antibody shows rare GFP+ cells in sections of Runx:GFP kidney marrow (brown; arrowheads; counterstained with hematoxilyn). Error bars show mean ± SEM. |
|
Time-Lapse Live-Imaging Sequence of CHT Colonization, Related to Figure 3 and Movie S2 Selected frames of time-lapse are shown (1 frame/2 min). First row frames are a merge of Runx:GFP+ HSPCs (green; second row) and kdrl:RFP+ ECs (red; third row in B). Times are hours:minutes post fertilization. Scale bars: 10 µm. (A) HSPC (arrowhead) extravasates by squeezing through endothelial wall. (B) ECs remodel around HSPC to form niche (broken circle). (C) After HSPC division the daughter cells undergo a 90° rotation (arrowheads). The upper cell becomes migratory and crawls out of niche. (D) A diagram summarizing the steps of HSPC lodgement in the perivascular niche. The times shown correspond to the time-lapse frames shown above. Other possible cell division decisions are shown. |
|
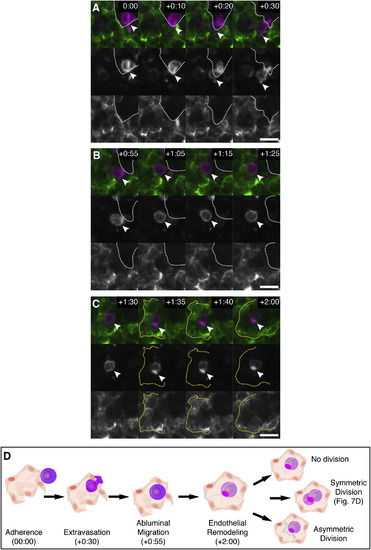
Time-Lapse Live Imaging of E11.5 FL Explant with Labeled HSPC and ECs, Related to Figure 4 (A) A time-lapse sequence from a FL lobe showing a c-kit+ HSPC (magenta) adhered to a CD31+ sinusoid (green). In ~30 min, the anterior of the cell protrudes (arrowhead) and extravasates to the abluminal side of the sinusoid (boundary marked by white dotted line). Luminal versus abluminal sides of the sinusoid were confirmed in single z slices of confocal stacks (data not shown). Times are hours:minutes after start of explant imaging. Scale bars: 10 µm. (B) After extravasation, the HSPC migrates on the abluminal side of the sinusoid. (C) ECs surround the HSPC as a rosette (yellow dotted line). (D) A diagram summarizing the steps of HSPC lodgement in the FL. The times shown correspond to the time-lapse frames shown above. A morphologically symmetric division is observed in Figure 4D. Other possible cell division decisions are shown. |
|
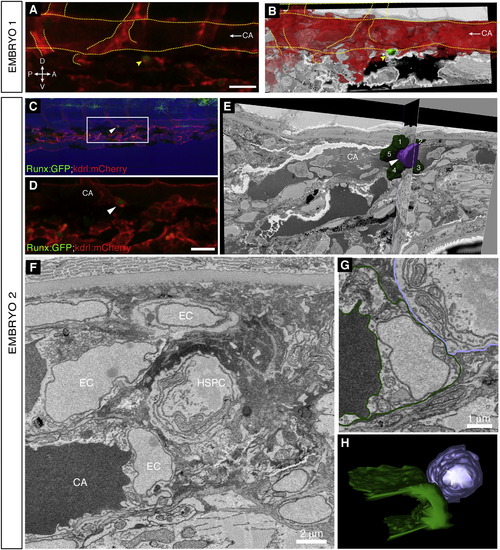
High-Resolution Electron Microscopy of Endogenous HSPC in the Perivascular Niche, Related to Figure 6 (A and B) Embryo 1 (same embryo as shown in Figure 6). Comparison of 3D projections from (A) confocal z-stack data and (B) serial EM scans to confirm the position of the lodged HSPC. (A) 3D rendered projection of z-stack (Imaris software) showing one lodged Runx:GFP+ HSPC (green; arrowhead) and kdrl:mCherry+ ECs (red). Scale bar: 25 µm. (B) Vessel lumen manually outlined, surface rendered, and shown in red (Imaris software). 3D projection overlaid on a single EM section. Rotation performed to match orientation of image in (A). Orientation: D (dorsal), V (ventral), A (anterior), P (posterior). CA (caudal artery). Circulation is toward the posterior (left; arrow). Major vessels outlined. (C?H) Embryo 2. A second independent example of serial block face EM sections on a lodged HSPC after time-lapse. (C) Last frame of CHT time-lapse (60 hpf). Arrowhead marks HSPC lodged > 6 hr. Runx:GFP green; kdrl:mCherry red; bright field blue. Anterior left, posterior right, dorsal top, ventral bottom. (D) Detail of region in (C) marked by box. The brightness of the GFP channel was adjusted independently of the mCherry channel to clearly show the position of the Runx:GFP+ HSPC. Scale bar: 25 µm. (E) Single section and orthogonal slice from serial block face EM scans. Lodged HSPC is purple (arrowhead), surrounding EC nuclei are green and numbered. Part of the HSPC is missing because it was not captured in serial EM sections. (F) High resolution EM section of HSPC in perivascular niche with cell nuclei labeled. (G) EM section with cell membranes outlined to show contact between EC (dark green) and HSPC (purple). (H) 3D modeling shows one side of the HSPC is in contact with the EC. For animated 3D reconstructions see Movie S7. |
|
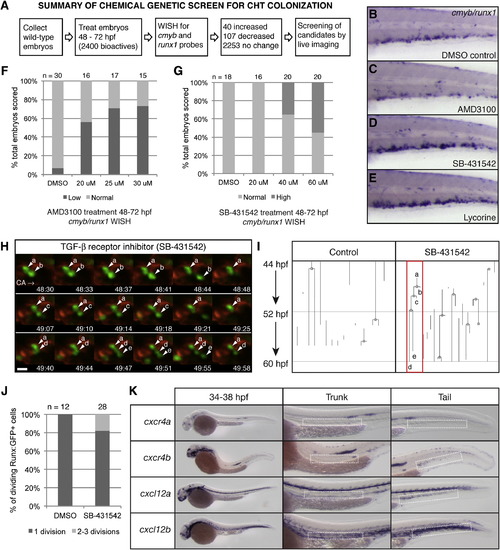
Chemical Genetic Screen to Identify Small-Molecule Regulators of CHT Niche Colonization, Related to Figure 7 (A) Overview of chemical screen. (B?E) whole mount in situ hybridization (WISH) of cmyb/runx1 probes in the CHT of 72 hpf embryos. Treatments from 48-72 hpf as indicated. (B) DMSO control with normal expression. (C?E) (C) Low expression after 25 µM AMD3100 treatment. High expression after treatment with (D) 40 µM SB-431542 or (E) 25 µM lycorine. (F and G) Wild-type AB embryos were treated from 48-72 hpf and scored as in (B-D). The percentage of total is shown. Number of embryos scored at top. (F) Dose-dependent decrease in cmyb/runx1 expression levels with increasing concentrations of CXCR4 antagonist AMD3100. The linear regression of Y (response) on X (dose) is significant: r2 = 0.3567, p = 7.84E-9. (G) Dose-dependent increase in cmyb/runx1 expression levels with increasing concentrations of TGF-β inhibitor, SB-431542. The linear regression of Y (response) on X (dose) is significant: r2 = 0.275081, p = 1.61E-6. (H) Selected frames from time-lapse movie of a Runx:GFP;kdrl:RFP double transgenic embryo treated with TGF-β inhibitor, SB-431542. In < 90 min, an HSPC (a) undergoes rounds of division and releases daughter cells (b,c,e) into circulation (daughter cell (d) was not released by the end of the movie; caudal artery, CA; circulation to posterior, right). Scale bar: 10 µm. (I) Lineage tree analysis of HSPCs tracked in parallel time-lapse movies, with and without SB-431542 treatment, from 44-60 hpf. One line is one continuously tracked HSPC from the time it enters the CHT field of view until it exits. Branch points mark clear cell divisions. The red box marks the lineage of the cells shown in (H). (J) Results from cell divisions scored in Runx:GFP+ time-lapse movies with DMSO or SB-431542 treatment, from 44-60 hpf. The percentage of total dividing cells scored. Total number of dividing cells scored at the top. Data collected from Runx:GFP+ time-lapse movies: DMSO treatment (11 embryos imaged; 42 Runx:GFP+ cells scored; 12/42 cells dividing; 0/12 cells dividing 2-3 times); SB-431542 treatment (15 embryos imaged; 79 Runx:GFP+ cells scored; 28/79 cells dividing; 5/28 cells dividing 2-3 times). (K) Whole mount in situ expression patterns of cxcr4a, cxcr4b, cxcl12a, and cxcl12b (stages range between 34-38 hpf). For each example a detail of the trunk and tail is shown, with the dorsal aorta and CHT, respectively, marked with dashed white boxes. All genes are expressed in the head and lateral line. cxcr4b but not cxcr4a is expressed in the CHT. cxcl12a and cxcl12b are expressed in the dorsal aorta and CHT. |
Reprinted from Cell, 160, Tamplin, O.J., Durand, E.M., Carr, L.A., Childs, S.J., Hagedorn, E.J., Li, P., Yzaguirre, A.D., Speck, N.A., Zon, L.I., Hematopoietic Stem Cell Arrival Triggers Dynamic Remodeling of the Perivascular Niche, 241-252, Copyright (2015) with permission from Elsevier. Full text @ Cell