- Title
-
A cdx4-sall4 regulatory module controls the transition from mesoderm formation to embryonic hematopoiesis
- Authors
- Paik, E.J., Mahony, S., White, R.M., Price, E.N., Dibiase, A., Dorjsuren, B., Mosimann, C., Davidson, A.J., Gifford, D., and Zon, L.I.
- Source
- Full text @ Stem Cell Reports
|
Cdx4 and Sall4 Bind to Each Other?s Genomic Locus (A) Cdx4-binding sites are enriched for the ATAAA motif. The most enriched motif was identified using MEME (Bailey and Elkan, 1994). (B) Gene track of the hox aa locus, showing Cdx4 binding at genomic regions along the x axis and the total number of reads on the y axis. (C) Gene track of the cdx4 locus, showing Cdx4 binding to its own locus. (D) Gene track of the sall4 locus, showing Cdx4 binding to sall4 locus. (E) WISH of sall4 mRNA expression level in 10-somite stage wild-type embryos and cdx4mo/cdx1amo. Scale bar = 200 Ám. (F) Sall4-binding sites are enriched for the ATTTGCAT motif, an Oct4 motif. (G) Gene track of the pou5f1 locus, showing Sall4 binding. (H) Gene track of sall4 and cdx4 loci, showing Sall4 binding. See also Figure S1. EXPRESSION / LABELING:
|
|
sall4 Cooperates with cdx4 in Zebrafish Hematopoiesis (A) gata1 WISH of 18-somite-stage wild-type or cdx4-/- embryos that were uninjected or sall4mo injected. sall4 knockdown in the cdx4-/- background leads to a decrease in gata1+ cells (note the high-powered view). (B) o-Dianisidine staining of 3 dpf wild-type or cdx4-/- embryos that were uninjected or sall4mo injected. cdx4-/-/sall4mo embryos have fewer hemoglobinized RBCs. Arrows point to the stained RBCs in the embryos. Scale bar, 200 Ám. See also Figure S3. EXPRESSION / LABELING:
PHENOTYPE:
|
|
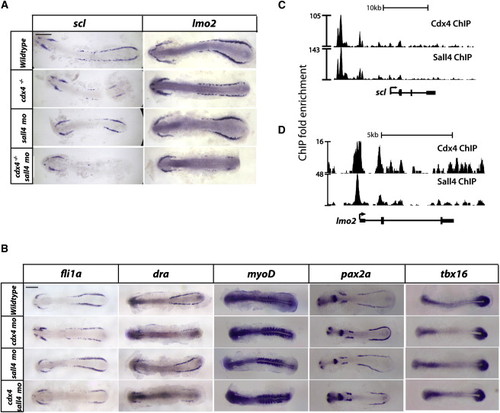
scl and lmo2 Are Responsible for the Loss of RBCs Seen in cdx4-/-;sall4mo Embryos. (A and B) scl (A) and lmo2 (B) WISH at the 10-somite stage wild-type or cdx4-/- embryos that are either uninjected or sall4mo injected. Embryos were flat-mounted, with the anterior end pointing to the left and the posterior end pointing to the right. (C) fli1a, draculin, pax2a, myoD, and tbx16 WISH in 10-somite stage wild-type, cdx4mo, sall4mo, cdx4mo/sall4mo embryos. All scale bars, 200 Ám. (D and E) Gene track of the scl (D) and lmo2 (E) loci, showing Cdx4 and Sall4 binding. See also Figure S4. EXPRESSION / LABELING:
|
|
Overexpression of scl and lmo2 Rescues the Loss of RBCs in the cdx4mo/sall4mo Embryos (A) DNA-mediated overexpression scheme using Tg(lcr:GFP) embryos that express GFP in globin+ RBCs. When the embryos reached the 18-somite stage, mCherry+ embryos were selected and their GFP expression and gata1 expression were examined. (B) cdx4mo/sall4mo/Tg(lcr:GFP) embryos that were control plasmid injected (top), ubi:scl injected (middle), or ubi:scl; ubi:lmo2 injected (bottom). The mCherry+ cells show the mosaicism. Note the GFP+ cells in the ICM (white arrow) in the bottom row. The numbers indicate the number of embryos with GFP signal in the ICM. (C) gata1 WISH of embryos shown in (B). The numbers indicate the number of embryos with gata1 signal in the ICM. All scale bars, 200 Ám. See also Figure S5. |