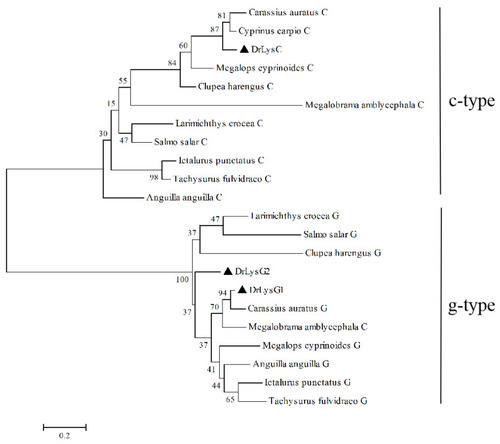
Phylogenetic analysis of zebrafish lysozymes and their homologs in fish. A phylogenetic tree was constructed by the neighbor-joining method using the Mega 4.0 software. The sequences of the lysozymes used in this analysis are as follows: DrLysC (NP_631919.1), Carassius auratus C (XP_026093809.1), Cyprinus carpio C (XP_018958317.1), Anguilla anguilla C (XP_035247404.1), Larimichthys crocea C (XP_019114157.2), Megalobrama amblycephala C (XP_048051439.1), Clupea harengus C (XP_012688691.1), Megalops cyprinoides C (XP_036378354.1), Ictalurus punctatus C (XP_017318497.1), Tachysurus fulvidraco C (XP_026996956.1), Salmo salar C (XP_014000972.1), DrLysG1 (NP_001002706.1), DrLysG2 (NP_001373416.1), Carassius auratus G (XP_026135699.1), Anguilla anguilla G (XP_035290251.1), Larimichthys crocea G (XP_010738712.1), Megalobrama amblycephala G (XP_048032707.1), Clupea harengus G (XP_031433526.1), Megalops cyprinoides G (XP_036404035.1), Ictalurus punctatus G (XP_017329515.1), Tachysurus fulvidraco G (XP_027010457.1), and Salmo salar G (XP_014031255.1). DrLysC, DrLysG1, and DrLysG2 are labeled by a triangle (▲). Numbers at nodes indicate bootstrap percentages (1000 replicates). The scale bar indicates evolutionary distance in base substitutions per site.
|

