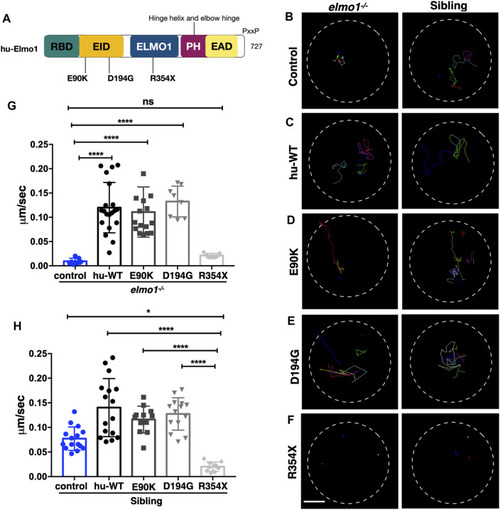
The zebrafish elmo1 mutant can serve as an in vivo model to verify the functions of human variants. (A) Schematic view of human ELMO1 protein conserved domains. Position of tested variants: p.E90K (E90K), p.D194G (D194G), and p.R354X (R354X) are indicated. (B?F) Track path of neutrophils expressing lyz:GFP control (B) or human ELMO1 (C?F) in the elmo1?/? and siblings recorded by live imaging at 3 dpf. Human wild-type (hu-WT) form (C), E90K (D), D194G (E), R354X (F). (G, H) Quantification of the migration speed of the control group and neutrophils expressing human ELMO1 variants in the elmo1?/? (7 cells of 3 larvae, 20 cells of 6 larvae, 16 cells of 6 larvae, 8 cells of 6 larvae, 7 cells of 3 larvae) (G) and sibling (15 cells of 3 larvae, 16 cells of 7 larvae, 12 cells of 8 larvae, 14 cells of 7 larvae, 10 cells of 5 larvae). (H). hu-WT, E90K, and D194G could efficiently rescue the migration speed in the elmo1 mutant compared with control. R354X failed to rescue the defects in elmo1?/? and even show a decreased migration speed in siblings. One-way ANOVA, ns: no significance, *p < 0.05, ****p < 0.001. Scale bar: 50 ?m (B?F).
|

